The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase
- PMID: 12186850
- PMCID: PMC2174009
- DOI: 10.1083/jcb.200205057
The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase
Abstract
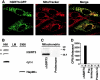
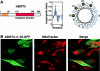
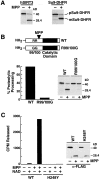
The yeast silent information regulator (Sir)2 protein links cellular metabolism and transcriptional silencing through its nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase activity. We report that mitochondria from mammalian cells contain intrinsic NAD-dependent deacetylase activity. This activity is inhibited by the NAD hydrolysis product nicotinamide, but not by trichostatin A, consistent with a class III deacetylase. We identify this deacetylase as the nuclear-encoded human Sir2 homologue hSIRT3, and show that hSIRT3 is located within the mitochondrial matrix. Mitochondrial import of hSIRT3 is dependent on an NH2-terminal amphipathic alpha-helix rich in basic residues. hSIRT3 is proteolytically processed in the mitochondrial matrix to a 28-kD product. This processing can be reconstituted in vitro with recombinant mitochondrial matrix processing peptidase (MPP) and is inhibited by mutation of arginines 99 and 100. The unprocessed form of hSIRT3 is enzymatically inactive and becomes fully activated in vitro after cleavage by MPP. These observations demonstrate the existence of a latent class III deacetylase that becomes catalytically activated upon import into the human mitochondria.
Figures


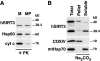
Similar articles
-
Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase.Nature. 2000 Feb 17;403(6771):795-800. doi: 10.1038/35001622. Nature. 2000. PMID: 10693811
-
Enzymatic activities of Sir2 and chromatin silencing.Curr Opin Cell Biol. 2001 Apr;13(2):232-8. doi: 10.1016/s0955-0674(00)00202-7. Curr Opin Cell Biol. 2001. PMID: 11248558 Review.
-
Role of NAD(+) in the deacetylase activity of the SIR2-like proteins.Biochem Biophys Res Commun. 2000 Nov 30;278(3):685-90. doi: 10.1006/bbrc.2000.3854. Biochem Biophys Res Commun. 2000. PMID: 11095969
-
Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition.Mol Cell. 2005 Feb 18;17(4):595-601. doi: 10.1016/j.molcel.2004.12.032. Mol Cell. 2005. PMID: 15721262
-
The Sir2 protein family: A novel deacetylase for gene silencing and more.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14030-2. doi: 10.1073/pnas.011506198. Proc Natl Acad Sci U S A. 2000. PMID: 11114164 Free PMC article. Review. No abstract available.
Cited by
-
Autophagy maintains ubiquitination-proteasomal degradation of Sirt3 to limit oxidative stress in K562 leukemia cells.Oncotarget. 2016 Jun 14;7(24):35692-35702. doi: 10.18632/oncotarget.9592. Oncotarget. 2016. PMID: 27232755 Free PMC article.
-
Post-translational modifications in mitochondria: protein signaling in the powerhouse.Cell Mol Life Sci. 2016 Nov;73(21):4063-73. doi: 10.1007/s00018-016-2280-4. Epub 2016 May 27. Cell Mol Life Sci. 2016. PMID: 27233499 Free PMC article. Review.
-
Mitochondrial SIRT4-type proteins in Caenorhabditis elegans and mammals interact with pyruvate carboxylase and other acetylated biotin-dependent carboxylases.Mitochondrion. 2013 Nov;13(6):705-20. doi: 10.1016/j.mito.2013.02.002. Epub 2013 Feb 21. Mitochondrion. 2013. PMID: 23438705 Free PMC article.
-
Do sirtuins promote mammalian longevity? A critical review on its relevance to the longevity effect induced by calorie restriction.Mol Cells. 2013 Jun;35(6):474-80. doi: 10.1007/s10059-013-0130-x. Epub 2013 May 8. Mol Cells. 2013. PMID: 23661364 Free PMC article. Review.
-
Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis.Sci Rep. 2012;2:425. doi: 10.1038/srep00425. Epub 2012 May 28. Sci Rep. 2012. PMID: 22645641 Free PMC article.
References
-
- Abe, Y., T. Shodai, T. Muto, K. Mihara, H. Torii, S. Nishikawa, T. Endo, and D. Kohda. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 100:551–560. - PubMed
-
- Afshar, G., and J.P. Murnane. 1999. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 234:161–168. - PubMed
-
- Arretz, M., H. Schneider, U. Wienhues, and W. Neupert. 1991. Processing of mitochondrial precursor proteins. Biomed. Biochim. Acta. 50:403–412. - PubMed
-
- Bernardi, P. 1999. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol. Rev. 79:1127–1155. - PubMed
-
- Bernardi, P., L. Scorrano, R. Colonna, V. Petronilli, and F. Di Lisa. 1999. Mitochondria and cell death. Mechanistic aspects and methodological issues. Eur. J. Biochem. 264:687–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

