Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence
- PMID: 12169668
- PMCID: PMC123205
- DOI: 10.1073/pnas.172380899
Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence
Abstract
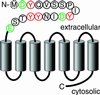
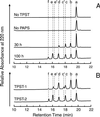
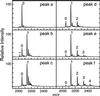
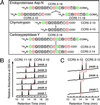
The CC-chemokine receptor 5 (CCR5) is the major coreceptor for the entry of macrophage-tropic (R5) HIV-1 strains into target cells. Posttranslational sulfation of tyrosine residues in the N-terminal tail of CCR5 is critical for high affinity interaction of the receptor with the HIV-1 envelope glycoprotein gp120 in complex with CD4. Here, we focused on defining precisely the sulfation pattern of the N terminus of CCR5 by using recombinant human tyrosylprotein sulfotransferases TPST-1 and TPST-2 to modify a synthetic peptide that corresponds to amino acids 2-18 of the receptor (CCR5 2-18). Analysis of the reaction products was made with a combination of reversed-phase HPLC, proteolytic cleavage, and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS). We found that CCR5 2-18 is sulfated by both TPST isoenzymes leading to a final product with four sulfotyrosine residues. Sulfates were added stepwise to the peptide producing specific intermediates with one, two, or three sulfotyrosines. The pattern of sulfation in these intermediates suggests that Tyr-14 and Tyr-15 are sulfated first, followed by Tyr-10, and finally Tyr-3. These results represent a detailed analysis of the multiple sulfation reaction of a peptide substrate by TPSTs and provide a structural basis for understanding the role of tyrosine sulfation of CCR5 in HIV-1 coreceptor and chemokine receptor function.
Figures

Similar articles
-
Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases.Biochemistry. 2008 Oct 28;47(43):11251-62. doi: 10.1021/bi800965m. Epub 2008 Oct 4. Biochemistry. 2008. PMID: 18834145 Free PMC article.
-
Pattern and temporal sequence of sulfation of CCR5 N-terminal peptides by tyrosylprotein sulfotransferase-2: an assessment of the effects of N-terminal residues.Biochemistry. 2009 Jun 16;48(23):5332-8. doi: 10.1021/bi900285c. Biochemistry. 2009. PMID: 19402700 Free PMC article.
-
Preparation and Analysis of N-Terminal Chemokine Receptor Sulfopeptides Using Tyrosylprotein Sulfotransferase Enzymes.Methods Enzymol. 2016;570:357-88. doi: 10.1016/bs.mie.2015.09.004. Epub 2015 Nov 14. Methods Enzymol. 2016. PMID: 26921955 Free PMC article.
-
The Synthesis of Sulfated CCR5 Peptide Surrogates and their Use to Study Receptor-Ligand Interactions.Protein Pept Lett. 2018;25(12):1124-1136. doi: 10.2174/0929866525666181101103834. Protein Pept Lett. 2018. PMID: 30381052 Review.
-
Determinants of tyrosylprotein sulfation coding and substrate specificity of tyrosylprotein sulfotransferases in metazoans.Chem Biol Interact. 2016 Nov 25;259(Pt A):17-22. doi: 10.1016/j.cbi.2016.04.006. Epub 2016 Apr 7. Chem Biol Interact. 2016. PMID: 27062897 Review.
Cited by
-
Direct mapping of tyrosine sulfation states in native peptides by nanopore.Nat Chem Biol. 2024 Sep 25. doi: 10.1038/s41589-024-01734-x. Online ahead of print. Nat Chem Biol. 2024. PMID: 39322788
-
Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.Sci Rep. 2017 Aug 18;7(1):8776. doi: 10.1038/s41598-017-07141-8. Sci Rep. 2017. PMID: 28821720 Free PMC article.
-
Synthetic DNA delivery by electroporation promotes robust in vivo sulfation of broadly neutralizing anti-HIV immunoadhesin eCD4-Ig.EBioMedicine. 2018 Sep;35:97-105. doi: 10.1016/j.ebiom.2018.08.027. Epub 2018 Aug 30. EBioMedicine. 2018. PMID: 30174283 Free PMC article.
-
SLC35B2 Acts in a Dual Role in the Host Sulfation Required for EV71 Infection.J Virol. 2022 May 11;96(9):e0204221. doi: 10.1128/jvi.02042-21. Epub 2022 Apr 14. J Virol. 2022. PMID: 35420441 Free PMC article.
-
Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12.Sci Signal. 2008 Sep 16;1(37):ra4. doi: 10.1126/scisignal.1160755. Sci Signal. 2008. PMID: 18799424 Free PMC article.
References
-
- Combadiere C., Ahuja, S. K., Tiffany, H. L. & Murphy, P. M. (1996) J. Leukocyte Biol. 60, 147-152. - PubMed
-
- Samson M., Labbe, O., Mollereau, C., Vassart, G. & Parmentier, M. (1996) Biochemistry 35, 3362-3367. - PubMed
-
- Raport C. J., Gosling, J., Schweickart, V. L., Gray, P. W. & Charo, I. F. (1996) J. Biol. Chem. 271, 17161-17166. - PubMed
-
- Gong W., Howard, O. M., Turpin, J. A., Grimm, M. C., Ueda, H., Gray, P. W., Raport, C. J., Oppenheim, J. J. & Wang, J. M. (1998) J. Biol. Chem. 273, 4289-4292. - PubMed
-
- Murphy P. M., Baggiolini, M., Charo, I. F., Hebert, C. A., Horuk, R., Matsushima, K., Miller, L. H., Oppenheim, J. J. & Power, C. A. (2000) Pharmacol. Rev. 52, 145-176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

