BRCA2 T2722R is a deleterious allele that causes exon skipping
- PMID: 12145750
- PMCID: PMC379197
- DOI: 10.1086/342192
BRCA2 T2722R is a deleterious allele that causes exon skipping
Erratum in
- Am J Hum Genet. 2003 Dec;73(6):1477
Abstract
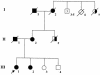
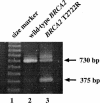
Patients with a strong family history of breast cancer are often counseled to receive genetic screening for BRCA1 and BRCA2 mutations, the strongest known predictors of breast cancer. A major limitation of genetic testing is the number of inconclusive results due to unclassified BRCA1 and BRCA2 sequence variants. Many known deleterious BRCA1 and BRCA2 mutations affect splicing, and these typically lie near intron/exon boundaries. However, there are also potential internal exonic mutations that disrupt functional exonic splicing enhancer (ESE) sequences, resulting in exon skipping. Using previously established sequence matrices for the scoring of putative ESE motifs, we have systematically examined several BRCA2 mutations for potential ESE disruption mutations. These predictions revealed that BRCA2 T2722R (8393C-->G), which segregates with affected individuals in a family with breast cancer, disrupts three potential ESE sites. Reverse-transcriptase polymerase chain reaction analysis confirms that this mutation causes exon skipping, leading to an out-of-frame fusion of BRCA2 exons 17 and 19. This represents the first BRCA2 missense mutation shown to be a predicted deleterious protein-truncating mutation and suggests a potentially useful method for determining the clinical significance of a subset of the many unclassified variants in BRCA1 and BRCA2.
Figures


Similar articles
-
Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene.J Med Genet. 2008 Jul;45(7):438-46. doi: 10.1136/jmg.2007.056895. Epub 2008 Apr 18. J Med Genet. 2008. PMID: 18424508
-
Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene.Hum Mutat. 2004 Nov;24(5):370-80. doi: 10.1002/humu.20087. Hum Mutat. 2004. PMID: 15459959
-
Functional classification of DNA variants by hybrid minigenes: Identification of 30 spliceogenic variants of BRCA2 exons 17 and 18.PLoS Genet. 2017 Mar 24;13(3):e1006691. doi: 10.1371/journal.pgen.1006691. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28339459 Free PMC article.
-
RNA analysis of eight BRCA1 and BRCA2 unclassified variants identified in breast/ovarian cancer families from Spain.Hum Mutat. 2003 Oct;22(4):337. doi: 10.1002/humu.9176. Hum Mutat. 2003. PMID: 12955719
-
The variants BRCA1 IVS6-1G>A and BRCA2 IVS15+1G>A lead to aberrant splicing of the transcripts.Breast Cancer Res Treat. 2009 Sep;117(2):461-5. doi: 10.1007/s10549-008-0154-7. Epub 2008 Aug 19. Breast Cancer Res Treat. 2009. PMID: 18712473
Cited by
-
Missense mutations of MLH1 and MSH2 genes detected in patients with gastrointestinal cancer are associated with exonic splicing enhancers and silencers.Oncol Lett. 2013 May;5(5):1710-1718. doi: 10.3892/ol.2013.1243. Epub 2013 Mar 11. Oncol Lett. 2013. PMID: 23760103 Free PMC article.
-
BRCA1 and BRCA2 germline mutation spectrum and frequencies in Belgian breast/ovarian cancer families.Br J Cancer. 2004 Mar 22;90(6):1244-51. doi: 10.1038/sj.bjc.6601656. Br J Cancer. 2004. PMID: 15026808 Free PMC article.
-
Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes.Nucleic Acids Res. 2005 Sep 7;33(16):5053-62. doi: 10.1093/nar/gki810. Print 2005. Nucleic Acids Res. 2005. PMID: 16147989 Free PMC article.
-
Genomic features defining exonic variants that modulate splicing.Genome Biol. 2010;11(2):R20. doi: 10.1186/gb-2010-11-2-r20. Epub 2010 Feb 16. Genome Biol. 2010. PMID: 20158892 Free PMC article.
-
BRCA1, BRCA2, TP53, and CDKN2A germline mutations in patients with breast cancer and cutaneous melanoma.Fam Cancer. 2007;6(4):453-61. doi: 10.1007/s10689-007-9143-y. Epub 2007 Jul 12. Fam Cancer. 2007. PMID: 17624602
References
Electronic-Database Information
-
- American Cancer Society: Breast Cancer Facts and Figures 2001–2002, http://www.cancer.org/eprise/main/docroot/stt/content/STT_1x_Breast_Canc...
-
- Breast Cancer Information Core, http://www.nhgri.nih.gov/Intramural_research/Lab_transfer/Bic/
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BRCA2 from mouse [accession number XM_124706], rat [accession number NM_031542], and chicken [accession number AY083934])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BRCA1 [MIM 113705] and BRCA2 [MIM 600185])
References
-
- Antoniou AC (2000) Risk models for familial ovarian and breast cancer. Genet Epidemiol 18:173–190 - PubMed
-
- Cartegni L, Chew S, Krainer A (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–300 - PubMed
-
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30:377–384 - PubMed
-
- Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, Ganguly A, Rebbeck T, Weber BL (1997) BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med 336:1409–1415 - PubMed
-
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343:692–695 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

