Transcriptional deregulation of the keratin 18 gene in human colon carcinoma cells results from an altered acetylation mechanism
- PMID: 12140315
- PMCID: PMC137086
- DOI: 10.1093/nar/gkf462
Transcriptional deregulation of the keratin 18 gene in human colon carcinoma cells results from an altered acetylation mechanism
Abstract
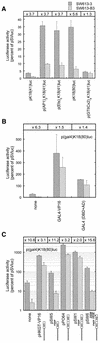
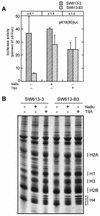
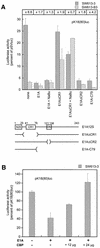
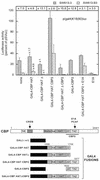
We are investigating the mechanism responsible for the overexpression of the keratin 18 (K18) gene in tumorigenic clones from the SW613-S human colon carcinoma cell line, as compared with non-tumorigenic clones. We have previously shown that this mechanism affects the minimal K18 promoter (TATA box and initiation site). We report here that treatment of the cells with histone deacetylase inhibitors stimulates the activity of the promoter in non-tumorigenic cells but has no effect in tumorigenic cells, resulting in a comparable activity of the promoter in both cell types. The adenovirus E1A protein inhibits the activity of the K18 promoter specifically in tumorigenic cells. This inhibition can be reversed by an excess of CBP protein. The conserved region 1 (CR1) of E1A, which is involved in the interaction with the CBP/p300 co-activators, is necessary to the inhibitory capacity of E1A. A 79 amino acid long N-terminal fragment of E1A, encompassing the two domains of E1A necessary and sufficient for binding to CBP (N-terminus and CR1), has the same differential inhibitory capacity on the K18 promoter as wild-type E1A. Forced recruitment of GAL4-CBP fusion proteins to the K18 promoter results in a greater stimulation of its activity in non-tumorigenic than in tumorigenic cells. The histone acetyltransferase activity of CBP is essential for this differential stimulation and the presence of the CBP2 domain greatly augments the activation capacity of the fusion protein. Chromatin immunoprecipitation experiments carried out with anti-acetylated histone antibodies showed no difference in the level of histone acetylation in the region of the K18 promoter between the two cell types. The structure of chromatin in the promoter region is similar in tumorigenic and non-tumorigenic cells, as determined by mapping of DNase I hypersensitive sites and probing the accessibility of the DNA to restriction endonucleases. From all these results we conclude that alteration of an acetylation mechanism involving the CBP (or p300) protein and acting on a non-histone substrate is responsible for the higher activity of the K18 promoter in tumorigenic cells of the SW613-S cell line.
Figures


Similar articles
-
Use of adenoviral E1A protein to analyze K18 promoter deregulation in colon carcinoma cells discloses a role for CtBP1 and BRCA1.BMC Mol Biol. 2005 Apr 14;6:8. doi: 10.1186/1471-2199-6-8. BMC Mol Biol. 2005. PMID: 15831101 Free PMC article.
-
Transcriptional mechanisms responsible for the overexpression of the keratin 18 gene in cells of a human colon carcinoma cell line.Exp Cell Res. 1999 Apr 10;248(1):243-59. doi: 10.1006/excr.1999.4402. Exp Cell Res. 1999. PMID: 10094831
-
Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region.Oncogene. 1997 Mar 6;14(9):1047-57. doi: 10.1038/sj.onc.1201002. Oncogene. 1997. PMID: 9070653
-
Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity.Mol Endocrinol. 2002 Apr;16(4):824-36. doi: 10.1210/mend.16.4.0811. Mol Endocrinol. 2002. PMID: 11923478
-
Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition.Cell Mol Life Sci. 2013 Nov;70(21):3989-4008. doi: 10.1007/s00018-012-1254-4. Epub 2013 Jan 11. Cell Mol Life Sci. 2013. PMID: 23307074 Free PMC article. Review.
Cited by
-
High frequency trans-splicing in a cell line producing spliced and polyadenylated RNA polymerase I transcripts from an rDNA-myc chimeric gene.Nucleic Acids Res. 2005 Apr 22;33(7):2332-42. doi: 10.1093/nar/gki530. Print 2005. Nucleic Acids Res. 2005. PMID: 15849319 Free PMC article.
-
Encapsulation of HaCaT Secretome for Enhanced Wound Healing Capacity on Human Dermal Fibroblasts.Mol Biotechnol. 2024 Jan;66(1):44-55. doi: 10.1007/s12033-023-00732-z. Epub 2023 Apr 4. Mol Biotechnol. 2024. PMID: 37016178
-
Hsa-miR-186-3p suppresses colon cancer progression by inhibiting KRT18/MAPK signaling pathway.Cell Cycle. 2022 Apr;21(7):741-753. doi: 10.1080/15384101.2021.2023305. Epub 2022 Mar 8. Cell Cycle. 2022. Retraction in: Cell Cycle. 2024 Jul 4:1. doi: 10.1080/15384101.2024.2370722 PMID: 35258413 Free PMC article. Retracted.
-
Use of adenoviral E1A protein to analyze K18 promoter deregulation in colon carcinoma cells discloses a role for CtBP1 and BRCA1.BMC Mol Biol. 2005 Apr 14;6:8. doi: 10.1186/1471-2199-6-8. BMC Mol Biol. 2005. PMID: 15831101 Free PMC article.
-
The relationship between keratin 18 and epithelial-derived tumors: as a diagnostic marker, prognostic marker, and its role in tumorigenesis.Front Oncol. 2024 Oct 22;14:1445978. doi: 10.3389/fonc.2024.1445978. eCollection 2024. Front Oncol. 2024. PMID: 39502314 Free PMC article. Review.
References
-
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. - PubMed
-
- Paranjape S.M., Kamakaka,R.T. and Kadonaga,J.T. (1994) Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem., 63, 265–297. - PubMed
-
- Wolffe A.P. (1992) New insights into chromatin function in transcriptional control. FASEB J., 6, 3354–3361. - PubMed
-
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. - PubMed
-
- Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

