Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities
- PMID: 12134021
- PMCID: PMC155159
- DOI: 10.1128/jvi.76.16.8161-8168.2002
Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities
Abstract
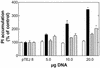
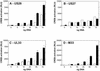
Cellular infection by cytomegalovirus (CMV) is associated with very early G-protein-mediated signal transduction and reprogramming of gene expression. Here we investigated the involvement of human CMV (HCMV)-encoded US27, US28, and UL33 receptors as well as murine CMV-encoded M33 transmembrane (7TM) receptors in host cell signaling mechanisms. HCMV-encoded US27 did not show any constitutive activity in any of the studied signaling pathways; in contrast, US28 and M33 displayed ligand-independent, constitutive signaling through the G protein q (Gq)/phospholipase C pathway. In addition, M33 and US28 also activated the transcription factor NF-kappaB as well as the cyclic AMP response element binding protein (CREB) in a ligand-independent, constitutive manner. The use of specific inhibitors indicated that the p38 mitogen-activated protein (MAP) kinase but not the extracellular signal-regulated kinase 1/2-MAP kinase pathway is involved in M33- and US28-mediated CREB activation but not NF-kappaB activation. Interestingly, UL33-the HCMV-encoded structural homologue of M33-was only marginally constitutively active in the Gq/phospholipase C turnover and CREB activation assays and did not show any constitutive activity in the NF-kappaB pathway, where M33 and US28 were highly active. Hence, CMVs appear to have conserved mechanisms for regulating host gene transcription, i.e., constitutive activation of certain kinases and transcription factors through the constitutive activities of 7TM proteins. These data, together with the previous identification of the incorporation of such proteins in the viral envelope, suggest that these proteins could be involved in the very early reprogramming of the host cell during viral infection.
Figures


Similar articles
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
Constitutive Signaling by the Human Cytomegalovirus G Protein Coupled Receptor Homologs US28 and UL33 Enables Trophoblast Migration In Vitro.Viruses. 2022 Feb 14;14(2):391. doi: 10.3390/v14020391. Viruses. 2022. PMID: 35215985 Free PMC article.
-
Functional analysis of the murine cytomegalovirus chemokine receptor homologue M33: ablation of constitutive signaling is associated with an attenuated phenotype in vivo.J Virol. 2008 Feb;82(4):1884-98. doi: 10.1128/JVI.02550-06. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057236 Free PMC article.
-
Human Cytomegalovirus US28: a functionally selective chemokine binding receptor.Infect Disord Drug Targets. 2009 Nov;9(5):548-56. doi: 10.2174/187152609789105696. Infect Disord Drug Targets. 2009. PMID: 19594424 Free PMC article. Review.
-
HCMV-encoded G-protein-coupled receptors as constitutively active modulators of cellular signaling networks.Trends Pharmacol Sci. 2006 Jan;27(1):56-63. doi: 10.1016/j.tips.2005.11.006. Epub 2005 Dec 13. Trends Pharmacol Sci. 2006. PMID: 16352349 Review.
Cited by
-
Molecular mechanisms deployed by virally encoded G protein-coupled receptors in human diseases.Annu Rev Pharmacol Toxicol. 2013;53:331-54. doi: 10.1146/annurev-pharmtox-010510-100608. Epub 2012 Oct 22. Annu Rev Pharmacol Toxicol. 2013. PMID: 23092247 Free PMC article. Review.
-
Do orphan G-protein-coupled receptors have ligand-independent functions? New insights from receptor heterodimers.EMBO Rep. 2006 Nov;7(11):1094-8. doi: 10.1038/sj.embor.7400838. EMBO Rep. 2006. PMID: 17077864 Free PMC article. Review.
-
The Human Cytomegalovirus US27 Gene Product Constitutively Activates Antioxidant Response Element-Mediated Transcription through Gβγ, Phosphoinositide 3-Kinase, and Nuclear Respiratory Factor 1.J Virol. 2018 Nov 12;92(23):e00644-18. doi: 10.1128/JVI.00644-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209167 Free PMC article.
-
The Mouse Cytomegalovirus G Protein-Coupled Receptor Homolog, M33, Coordinates Key Features of In Vivo Infection via Distinct Components of Its Signaling Repertoire.J Virol. 2022 Feb 23;96(4):e0186721. doi: 10.1128/JVI.01867-21. Epub 2021 Dec 8. J Virol. 2022. PMID: 34878888 Free PMC article.
-
Polymorphisms within human cytomegalovirus chemokine (UL146/UL147) and cytokine receptor genes (UL144) are not predictive of sequelae in congenitally infected children.Virology. 2008 Aug 15;378(1):86-96. doi: 10.1016/j.virol.2008.05.002. Epub 2008 Jun 16. Virology. 2008. PMID: 18556037 Free PMC article.
References
-
- Abubakar, S., I. Boldogh, and T. Albrecht. 1990. Human cytomegalovirus stimulates arachidonic acid metabolism through pathways that are affected by inhibitors of phospholiphase A2 and protein kinase C. Biochem. Biophys. Res. Commun. 166:953-959. - PubMed
-
- Alford, C. A., and W. J. Britt. 1995. Cytomegaloviruses, p. 2493-2534. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, New York, N.Y.
-
- Ambinder, R. F., K. D. Robertson, and Q. Tao. 1999. DNA methylation and the Epstein-Barr virus. Semin. Cancer Biol. 9:369-375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

