Identification of an upstream sequence element required for vesicular stomatitis virus mRNA transcription
- PMID: 12097577
- PMCID: PMC136381
- DOI: 10.1128/jvi.76.15.7632-7641.2002
Identification of an upstream sequence element required for vesicular stomatitis virus mRNA transcription
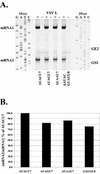
Abstract
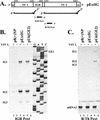
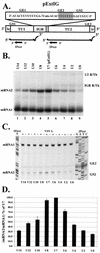
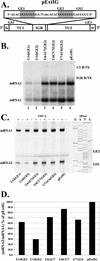
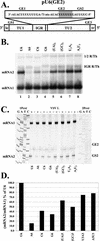
Vesicular stomatitis virus (VSV), the prototypic rhabdovirus, has a nonsegmented negative-sense RNA genome with five genes flanked by 3' leader and 5' trailer sequences. Transcription of VSV mRNAs is obligatorily sequential, starting from a single 3' polymerase entry site, and termination of an upstream mRNA is essential for transcription of a downstream gene. cis-acting signals for transcription of VSV mRNAs are present within the leader region, at the leader-N junction, and at the internal gene junctions. The gene junctions of VSV consist of a conserved 23-nucleotide region that includes the gene end sequence of the upstream gene, 3'-AUACU7-5', a nontranscribed intergenic dinucleotide, 3'-G/CA-5', and the gene start sequence, 3'-UUGUCNNUAG-5', at the beginning of the gene immediately downstream. Previous work has shown that the gene end sequence and intergenic region are sufficient to signal polyadenylation and termination of VSV transcripts. Mutagenesis of the gene start sequence has determined the importance of this region in the processes of initiation and 5'-end modification of mRNAs. However, because the gene end sequence is positioned directly upstream of the gene start sequence in the gene junction, and because of the requirement for termination of the upstream gene prior to transcription of the downstream gene, it has not been possible to investigate whether the gene end sequence contributes to transcription of the downstream gene. In this study, we inserted an additional gene end sequence upstream of the gene junction in a subgenomic replicon of VSV, which extended the intergenic region from 2 to 88 nucleotides. This duplication of termination signals allowed us to separate the signals required for termination from those required for initiation. We investigated the effect that the upstream gene end sequences had on downstream mRNA transcription. Our data show that the U7 tract of the upstream gene end sequence is necessary for optimal transcription of the downstream gene, independent of its role in termination of the upstream gene. Altering the sequence or changing the length of the U tract directly upstream of the gene start sequence significantly decreased transcription of the downstream gene. These results show that the U tract is a multifunctional region that is required not only for polyadenylation and termination of the upstream mRNA but also for efficient transcription of the downstream gene.
Figures

Similar articles
-
The VSV polymerase can initiate at mRNA start sites located either up or downstream of a transcription termination signal but size of the intervening intergenic region affects efficiency of initiation.Virology. 2008 May 10;374(2):361-70. doi: 10.1016/j.virol.2007.12.023. Epub 2008 Jan 31. Virology. 2008. PMID: 18241907 Free PMC article.
-
Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts.J Virol. 1997 Mar;71(3):2127-37. doi: 10.1128/JVI.71.3.2127-2137.1997. J Virol. 1997. PMID: 9032346 Free PMC article.
-
Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions.J Virol. 1998 Mar;72(3):1805-13. doi: 10.1128/JVI.72.3.1805-1813.1998. J Virol. 1998. PMID: 9499031 Free PMC article.
-
Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus.Biochim Biophys Acta. 2002 Sep 13;1577(2):337-53. doi: 10.1016/s0167-4781(02)00462-1. Biochim Biophys Acta. 2002. PMID: 12213662 Review.
-
Transcription and replication of nonsegmented negative-strand RNA viruses.Curr Top Microbiol Immunol. 2004;283:61-119. doi: 10.1007/978-3-662-06099-5_3. Curr Top Microbiol Immunol. 2004. PMID: 15298168 Review.
Cited by
-
Analysis of the highly diverse gene borders in Ebola virus reveals a distinct mechanism of transcriptional regulation.J Virol. 2014 Nov;88(21):12558-71. doi: 10.1128/JVI.01863-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142600 Free PMC article.
-
Sendai Virus and a Unified Model of Mononegavirus RNA Synthesis.Viruses. 2021 Dec 9;13(12):2466. doi: 10.3390/v13122466. Viruses. 2021. PMID: 34960735 Free PMC article. Review.
-
Role of intergenic sequences in newcastle disease virus RNA transcription and pathogenesis.J Virol. 2008 Feb;82(3):1323-31. doi: 10.1128/JVI.01989-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032502 Free PMC article.
-
How does the polymerase of non-segmented negative strand RNA viruses commit to transcription or genome replication?J Virol. 2024 Aug 20;98(8):e0033224. doi: 10.1128/jvi.00332-24. Epub 2024 Jul 30. J Virol. 2024. PMID: 39078194 Review.
-
Conserved aspartic acid 233 and alanine 231 are not required for poliovirus polymerase function in replicons.Virol J. 2007 Mar 12;4:28. doi: 10.1186/1743-422X-4-28. Virol J. 2007. PMID: 17352827 Free PMC article.
References
-
- Banerjee, A. D., G. Abraham, and R. J. Colonno. 1977. Vesicular stomatitis virus: mode of transcription. J. Gen. Virol. 34:1-8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

