Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR
- PMID: 12097568
- PMCID: PMC136374
- DOI: 10.1128/jvi.76.15.7554-7559.2002
Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR
Abstract
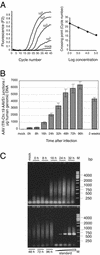
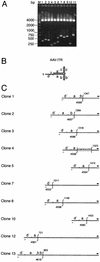
Adeno-associated virus type 2 (AAV-2) integrates specifically into a site on human chromosome 19 (chr-19) called AAVS1. To study the kinetics and frequency of chr-19-specific integration after AAV infection, we developed a rapid, sensitive, and quantitative real-time PCR assay for AAV inverted terminal repeat-chr-19-specific junctions. Despite the known variability of junction sites, conditions were established that ensured reliable quantification of integration rates within hours after AAV infection. The overall integration frequency was calculated to peak at between 10 and 20% of AAV-infected, unselected HeLa cells. At least 1 in 1,000 infectious AAV-2 particles was found to integrate site specifically up to day 4 postinfection in the absence of selection. Chromosomal breakpoints within AAVS1 agreed with those found in latently infected clonal cell lines and transgenic animals. Use of this quantitative real-time PCR will greatly facilitate the study of the early steps of wild-type and recombinant AAV vector integration.
Figures


Similar articles
-
Adeno-associated virus integrates site-specifically into human chromosome 19 in either orientation and with equal kinetics and frequency.J Gen Virol. 2003 Jan;84(Pt 1):133-137. doi: 10.1099/vir.0.18726-0. J Gen Virol. 2003. PMID: 12533709
-
Packaging of human chromosome 19-specific adeno-associated virus (AAV) integration sites in AAV virions during AAV wild-type and recombinant AAV vector production.J Virol. 2003 Apr;77(8):4881-7. doi: 10.1128/jvi.77.8.4881-4887.2003. J Virol. 2003. PMID: 12663794 Free PMC article.
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
Site-specific integration by adeno-associated virus.Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11288-94. doi: 10.1073/pnas.93.21.11288. Proc Natl Acad Sci U S A. 1996. PMID: 8876128 Free PMC article. Review.
-
Integration of adeno-associated virus (AAV) and recombinant AAV vectors.Annu Rev Genet. 2004;38:819-45. doi: 10.1146/annurev.genet.37.110801.143717. Annu Rev Genet. 2004. PMID: 15568995 Review.
Cited by
-
Adeno-associated virus type 2 p5 promoter: a rep-regulated DNA switch element functioning in transcription, replication, and site-specific integration.J Virol. 2007 Apr;81(8):3721-30. doi: 10.1128/JVI.02693-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267512 Free PMC article.
-
Evidence of chromosomal integration of AAV DNA in human testis tissue.Virus Genes. 2004 Jan;28(1):61-9. doi: 10.1023/B:VIRU.0000012264.54212.f5. Virus Genes. 2004. PMID: 14739652
-
Adeno-associated virus site-specific integration is regulated by TRP-185.J Virol. 2007 Feb;81(4):1990-2001. doi: 10.1128/JVI.02014-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151120 Free PMC article.
-
Adeno-associated virus site-specific integration and AAVS1 disruption.J Virol. 2004 Aug;78(15):7874-82. doi: 10.1128/JVI.78.15.7874-7882.2004. J Virol. 2004. PMID: 15254160 Free PMC article.
-
Targeted chromosomal insertion of large DNA into the human genome by a fiber-modified high-capacity adenovirus-based vector system.PLoS One. 2008 Aug 29;3(8):e3084. doi: 10.1371/journal.pone.0003084. PLoS One. 2008. PMID: 18769728 Free PMC article.
References
-
- Berns, K. I. 1996. Parvoviridae: the viruses and their replication, p. 2173-2197. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
-
- Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum. Gene Ther. 9:2745-2760. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

