Identification of gene function by cyclical packaging rescue of retroviral cDNA libraries
- PMID: 12084928
- PMCID: PMC124385
- DOI: 10.1073/pnas.132274799
Identification of gene function by cyclical packaging rescue of retroviral cDNA libraries
Abstract
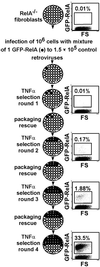
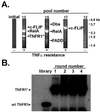
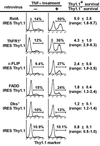
Genes regulating responses in mammalian cells are often difficult to identify by functional cloning strategies limited to a single round of selection. Here we describe a strategy, cyclical packaging rescue (CPR), which allows rapid recovery and retransmission of retroviral cDNA libraries. CPR can be used not only with immortalized cell lines such as fibroblasts and Jurkat T cells, but also with primary B lymphocytes, which can be maintained only in short-term cultures. CPR allows for multiple rounds of selection and enrichment to identify cDNAs regulating responses in mammalian cells. Using CPR, five cDNAs were functionally cloned, which conferred protection against tumor necrosis factor alpha (TNFalpha)-induced apoptosis in RelA(-/-) fibroblasts. Three of the genes, RelA, cellular FLICE-like inhibitory protein (c-FLIP), and a dominant-negative mutant of TNF receptor 1 arising through CPR afforded strong protection against apoptosis. Two of the genes identified, Dbs and Fas-associated death domain protein (FADD), previously identified as a proapoptotic molecule, afforded partial protection against TNFalpha-induced apoptosis. These results suggest that CPR is a versatile method that permits functional identification of both wild-type and dominant-negative gene products that regulate cellular responses.
Figures

Similar articles
-
[Screening of drug resistent gene by cyclical packaging rescue of hepatocellular carcinoma retroviral cDNA libraries].Sheng Wu Gong Cheng Xue Bao. 2016 Feb;32(2):204-11. Sheng Wu Gong Cheng Xue Bao. 2016. PMID: 27382770 Chinese.
-
Expression cloning of oncogenes by retroviral transfer of cDNA libraries.Mol Cell Biol. 1995 Feb;15(2):704-10. doi: 10.1128/MCB.15.2.704. Mol Cell Biol. 1995. PMID: 7823939 Free PMC article.
-
FADD deficiency sensitises Jurkat T cells to TNF-alpha-dependent necrosis during activation-induced cell death.FEBS Lett. 2005 Nov 21;579(28):6465-72. doi: 10.1016/j.febslet.2005.10.041. Epub 2005 Nov 2. FEBS Lett. 2005. PMID: 16289096
-
Dissection of signaling pathways and cloning of new signal transducers in tyrosine kinase-induced pathways by genetic selection.Leukemia. 1998 Dec;12(12):1858-65. doi: 10.1038/sj.leu.2401231. Leukemia. 1998. PMID: 9844916 Review.
-
Crosstalk between apoptosis, necrosis and autophagy.Biochim Biophys Acta. 2013 Dec;1833(12):3448-3459. doi: 10.1016/j.bbamcr.2013.06.001. Epub 2013 Jun 13. Biochim Biophys Acta. 2013. PMID: 23770045 Review.
Cited by
-
Fas-associated death domain protein interacts with methyl-CpG binding domain protein 4: a potential link between genome surveillance and apoptosis.Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5211-6. doi: 10.1073/pnas.0431215100. Epub 2003 Apr 17. Proc Natl Acad Sci U S A. 2003. PMID: 12702765 Free PMC article.
-
Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation.J Immunol. 2007 Nov 15;179(10):6808-19. doi: 10.4049/jimmunol.179.10.6808. J Immunol. 2007. PMID: 17982071 Free PMC article.
-
iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS.Science. 2012 Jan 13;335(6065):229-32. doi: 10.1126/science.1214448. Science. 2012. PMID: 22246778 Free PMC article.
-
Cell entry of enveloped viruses.Adv Genet. 2011;73:121-83. doi: 10.1016/B978-0-12-380860-8.00004-5. Adv Genet. 2011. PMID: 21310296 Free PMC article. Review.
-
Spontaneous mutation of the Dock2 gene in Irf5-/- mice complicates interpretation of type I interferon production and antibody responses.Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):E898-904. doi: 10.1073/pnas.1118155109. Epub 2012 Mar 19. Proc Natl Acad Sci U S A. 2012. PMID: 22431588 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

