Rad23 promotes the targeting of proteolytic substrates to the proteasome
- PMID: 12052895
- PMCID: PMC133919
- DOI: 10.1128/MCB.22.13.4902-4913.2002
Rad23 promotes the targeting of proteolytic substrates to the proteasome
Abstract
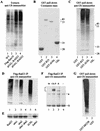
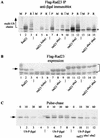
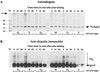
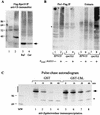
Rad23 contains a ubiquitin-like domain (UbL(R23)) that interacts with catalytically active proteasomes and two ubiquitin (Ub)-associated (UBA) sequences that bind Ub. The UBA domains can bind Ub in vitro, although the significance of this interaction in vivo is poorly understood. Rad23 can interfere with the assembly of multi-Ub chains in vitro, and high-level expression caused stabilization of proteolytic substrates in vivo. We report here that Rad23 interacts with ubiquitinated cellular proteins through the synergistic action of its UBA domains. Rad23 plays an overlapping role with Rpn10, a proteasome-associated multi-Ub chain binding protein. Mutations in the UBA domains prevent efficient interaction with ubiquitinated proteins and result in poor suppression of the growth and proteolytic defects of a rad23 Delta rpn10 Delta mutant. High-level expression of Rad23 revealed, for the first time, an interaction between ubiquitinated proteins and the proteasome. This increase was not observed in rpn10 Delta mutants, suggesting that Rpn10 participates in the recognition of proteolytic substrates that are delivered by Rad23. Overexpression of UbL(R23) caused stabilization of a model substrate, indicating that an unregulated UbL(R23)-proteasome interaction can interfere with the efficient delivery of proteolytic substrates by Rad23. Because the suppression of a rad23 Delta rpn10 Delta mutant phenotype required both UbL(R23) and UBA domains, our findings support the hypothesis that Rad23 encodes a novel regulatory factor that translocates ubiquitinated substrates to the proteasome.
Figures


Similar articles
-
Investigating the importance of proteasome-interaction for Rad23 function.Curr Genet. 2003 Jan;42(4):199-208. doi: 10.1007/s00294-002-0350-7. Epub 2002 Dec 13. Curr Genet. 2003. PMID: 12589471
-
Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain.J Mol Biol. 2014 Dec 12;426(24):4049-4060. doi: 10.1016/j.jmb.2014.10.004. Epub 2014 Oct 13. J Mol Biol. 2014. PMID: 25311859 Free PMC article.
-
Proteasome subunit Rpn1 binds ubiquitin-like protein domains.Nat Cell Biol. 2002 Sep;4(9):725-30. doi: 10.1038/ncb845. Nat Cell Biol. 2002. PMID: 12198498
-
Rad23 and Rpn10: perennial wallflowers join the melee.Trends Biochem Sci. 2004 Dec;29(12):637-40. doi: 10.1016/j.tibs.2004.10.008. Trends Biochem Sci. 2004. PMID: 15544949 Review.
-
The moonlighting of RAD23 in DNA repair and protein degradation.Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194925. doi: 10.1016/j.bbagrm.2023.194925. Epub 2023 Feb 28. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36863450 Review.
Cited by
-
ATP binding by proteasomal ATPases regulates cellular assembly and substrate-induced functions of the 26 S proteasome.J Biol Chem. 2013 Feb 1;288(5):3334-45. doi: 10.1074/jbc.M112.424788. Epub 2012 Dec 4. J Biol Chem. 2013. PMID: 23212908 Free PMC article.
-
The ubiquitin-associated (UBA) 1 domain of Schizosaccharomyces pombe Rhp23 is essential for the recognition of ubiquitin-proteasome system substrates both in vitro and in vivo.J Biol Chem. 2012 Dec 7;287(50):42344-51. doi: 10.1074/jbc.M112.419838. Epub 2012 Oct 4. J Biol Chem. 2012. PMID: 23038266 Free PMC article.
-
Such small hands: the roles of centrins/caltractins in the centriole and in genome maintenance.Cell Mol Life Sci. 2012 Sep;69(18):2979-97. doi: 10.1007/s00018-012-0961-1. Epub 2012 Mar 30. Cell Mol Life Sci. 2012. PMID: 22460578 Free PMC article. Review.
-
The Snf1 kinase and proteasome-associated Rad23 regulate UV-responsive gene expression.EMBO J. 2009 Oct 7;28(19):2919-31. doi: 10.1038/emboj.2009.229. Epub 2009 Aug 13. EMBO J. 2009. PMID: 19680226 Free PMC article.
-
Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin.J Mol Biol. 2007 May 25;369(1):168-76. doi: 10.1016/j.jmb.2007.03.008. Epub 2007 Mar 12. J Mol Biol. 2007. PMID: 17408689 Free PMC article.
References
-
- Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugawawa, J. Kondoh, Y. Okhuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276:18665-18672. - PubMed
-
- Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. - PubMed
-
- Beal, R. E., D. Toscano-Cantaffa, P. Young, M. Rechsteiner, and C. M. Pickart. 1998. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry 37:2925-2934. - PubMed
-
- Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J. Mol. Biol. 313:955-963. - PubMed
-
- Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
