The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I
- PMID: 12052259
- PMCID: PMC115839
- DOI: 10.1186/1471-2091-3-13
The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I
Abstract
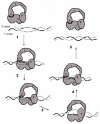
Background: Escherichia coli DNA topoisomerase I binds three Zn(II) with three tetracysteine motifs which, together with the 14 kDa C-terminal region, form a 30 kDa DNA binding domain (ZD domain). The 67 kDa N-terminal domain (Top67) has the active site tyrosine for DNA cleavage but cannot relax negatively supercoiled DNA. We analyzed the role of the ZD domain in the enzyme mechanism.
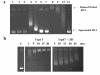
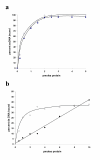
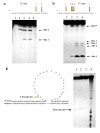
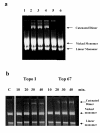
Results: Addition of purified ZD domain to Top67 partially restored the relaxation activity, demonstrating that covalent linkage between the two domains is not necessary for removal of negative supercoils from DNA. The two domains had similar affinities to ssDNA. However, only Top67 could bind dsDNA with high affinity. DNA cleavage assays showed that the Top67 had the same sequence and structure selectivity for DNA cleavage as the intact enzyme. DNA rejoining also did not require the presence of the ZD domain.
Conclusions: We propose that during relaxation of negatively supercoiled DNA, Top67 by itself can position the active site tyrosine near the junction of double-stranded and single-stranded DNA for cleavage. However, the interaction of the ZD domain with the passing single-strand of DNA, coupled with enzyme conformational change, is needed for removal of negative supercoils.
Figures


Similar articles
-
Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I.Nucleic Acids Res. 2015 Dec 15;43(22):11031-46. doi: 10.1093/nar/gkv1073. Epub 2015 Oct 20. Nucleic Acids Res. 2015. PMID: 26490962 Free PMC article.
-
The Zn(II) binding motifs of E. coli DNA topoisomerase I is part of a high-affinity DNA binding domain.Biochem Biophys Res Commun. 1998 Oct 20;251(2):509-14. doi: 10.1006/bbrc.1998.9500. Biochem Biophys Res Commun. 1998. PMID: 9792804
-
Iron inhibits Escherichia coli topoisomerase I activity by targeting the first two zinc-binding sites in the C-terminal domain.Protein Sci. 2014 Nov;23(11):1619-28. doi: 10.1002/pro.2542. Epub 2014 Sep 13. Protein Sci. 2014. PMID: 25176012 Free PMC article.
-
The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding.J Biol Chem. 1994 Mar 25;269(12):9052-9. J Biol Chem. 1994. PMID: 7510701
-
Bacterial and archeal type I topoisomerases.Biochim Biophys Acta. 1998 Oct 1;1400(1-3):19-27. doi: 10.1016/s0167-4781(98)00125-0. Biochim Biophys Acta. 1998. PMID: 9748482 Review.
Cited by
-
Mechanism of Type IA Topoisomerases.Molecules. 2020 Oct 17;25(20):4769. doi: 10.3390/molecules25204769. Molecules. 2020. PMID: 33080770 Free PMC article. Review.
-
Metal ion and inter-domain interactions as functional networks in E. coli topoisomerase I.Gene. 2013 Jul 25;524(2):253-60. doi: 10.1016/j.gene.2013.04.008. Epub 2013 Apr 20. Gene. 2013. PMID: 23612251 Free PMC article.
-
What's on the Other Side of the Gate: A Structural Perspective on DNA Gate Opening of Type IA and IIA DNA Topoisomerases.Int J Mol Sci. 2023 Feb 16;24(4):3986. doi: 10.3390/ijms24043986. Int J Mol Sci. 2023. PMID: 36835394 Free PMC article. Review.
-
C-terminal lysine repeats in Streptomyces topoisomerase I stabilize the enzyme-DNA complex and confer high enzyme processivity.Nucleic Acids Res. 2017 Nov 16;45(20):11908-11924. doi: 10.1093/nar/gkx827. Nucleic Acids Res. 2017. PMID: 28981718 Free PMC article.
-
Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage.Nucleic Acids Res. 2013 Aug;41(15):7462-71. doi: 10.1093/nar/gkt506. Epub 2013 Jun 14. Nucleic Acids Res. 2013. PMID: 23771144 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

