Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast
- PMID: 12034844
- PMCID: PMC117174
- DOI: 10.1093/nar/30.11.2546
Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast
Abstract
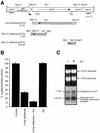
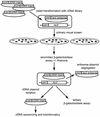
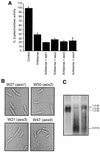
Specific gene silencing has been demonstrated in a number of organisms by the introduction of antisense RNA. Mutagenesis of host-encoded factors has begun to unravel the mechanism of several forms of RNA-mediated gene silencing and has suggested that it may have been conserved through evolution. This has led to the identification of certain host genes, which, when mutated, abrogate this phenomenon. Conversely, the identification of other factors that, when co-expressed or overexpressed, can enhance gene inhibition is equally important for both elucidating the mechanism of this process and enhancing gene silencing in recalcitrant systems. We have taken such a dominant genetic approach to identify several host-encoded factors that dramatically enhance target gene silencing when co-expressed with antisense RNA in fission yeast. The transcription factor thi1 and, surprisingly, the ATP-dependent RNA helicase ded1 were initially shown to enhance gene silencing in this system. Additionally, screening of a Schizosaccharomyces pombe cDNA library identified four novel antisense-enhancing sequences (aes factors) all of which are homologous to genes encoding proteins with natural affinities for nucleic acids. These findings demonstrate the utility of this strategy in identifying host-encoded factors that can modulate gene silencing when co-expressed with antisense RNA and possibly other forms of gene-silencing activators.
Figures



Similar articles
-
Double-stranded RNA-mediated gene silencing in fission yeast.Nucleic Acids Res. 2003 Aug 1;31(15):4481-9. doi: 10.1093/nar/gkg484. Nucleic Acids Res. 2003. PMID: 12888508 Free PMC article.
-
The ste13+ gene encoding a putative RNA helicase is essential for nitrogen starvation-induced G1 arrest and initiation of sexual development in the fission yeast Schizosaccharomyces pombe.Mol Gen Genet. 1994 Sep 1;244(5):456-64. doi: 10.1007/BF00583896. Mol Gen Genet. 1994. PMID: 8078473
-
Gene regulation by antisense RNA in the fission yeast Schizosaccharomyces pombe.Mol Gen Genet. 1995 Aug 21;248(3):293-300. doi: 10.1007/BF02191596. Mol Gen Genet. 1995. PMID: 7565591
-
RNA interference pathways in fungi: mechanisms and functions.Annu Rev Microbiol. 2012;66:305-23. doi: 10.1146/annurev-micro-092611-150138. Epub 2012 Jun 28. Annu Rev Microbiol. 2012. PMID: 22746336 Free PMC article. Review.
-
Mechanisms of gene regulation by endogenous and artificially introduced antisense RNA.Biochem Soc Trans. 1992 Nov;20(4):750-4. doi: 10.1042/bst0200750. Biochem Soc Trans. 1992. PMID: 1487056 Review. No abstract available.
Cited by
-
Double-stranded RNA-mediated gene silencing in fission yeast.Nucleic Acids Res. 2003 Aug 1;31(15):4481-9. doi: 10.1093/nar/gkg484. Nucleic Acids Res. 2003. PMID: 12888508 Free PMC article.
-
DEAD-box helicase proteins disrupt RNA tertiary structure through helix capture.PLoS Biol. 2014 Oct 28;12(10):e1001981. doi: 10.1371/journal.pbio.1001981. eCollection 2014 Oct. PLoS Biol. 2014. PMID: 25350280 Free PMC article.
-
Open reading frames provide a rich pool of potential natural antisense transcripts in fungal genomes.Nucleic Acids Res. 2005 Sep 7;33(16):5034-44. doi: 10.1093/nar/gki804. Print 2005. Nucleic Acids Res. 2005. PMID: 16147987 Free PMC article.
-
Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.Proc Natl Acad Sci U S A. 2004 Nov 23;101(47):16431-6. doi: 10.1073/pnas.0407371101. Epub 2004 Nov 15. Proc Natl Acad Sci U S A. 2004. PMID: 15545603 Free PMC article.
-
The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex.Mol Cell. 2011 Sep 16;43(6):962-72. doi: 10.1016/j.molcel.2011.08.008. Mol Cell. 2011. PMID: 21925384 Free PMC article.
References
-
- Murray J. and Crockett,N. (1992) Antisense RNA; an overview. In Murray,J. (ed.), Antisense RNA and DNA. Wiley-Liss, New York, NY, pp. 1–49.
-
- Sczakiel G. (1997) The design of antisense RNA. Antisense Nucleic Acid Drug Dev., 7, 439–444. - PubMed
-
- Fire A., Xu,S., Montgomery,M., Kostas,S., Driver,S. and Mello,C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.Nature, 391, 806–811. - PubMed
-
- Denhardt D. (1992) Mechanism of action of antisense RNA. Ann. N. Y. Acad. Sci., 662, 70–76. - PubMed
-
- Raponi M., Atkins,D., Dawes,I. and Arndt,G. (2000) The influence of antisense gene location on target gene regulation in the fission yeast Schizosaccharomyces pombe.Antisense Nucleic Acid Drug Dev., 10, 29–34. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

