Bacillus subtilis bacteriophage SPP1 hexameric DNA helicase, G40P, interacts with forked DNA
- PMID: 12034814
- PMCID: PMC117191
- DOI: 10.1093/nar/30.11.2280
Bacillus subtilis bacteriophage SPP1 hexameric DNA helicase, G40P, interacts with forked DNA
Abstract
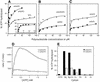
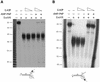
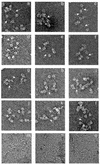
SPP1-encoded replicative DNA helicase gene 40 product (G40P) is an essential product for phage replication. Hexameric G40P, in the presence of AMP-PNP, preferentially binds unstructured single-stranded (ss)DNA in a sequence-independent manner. The efficiency of ssDNA binding, nucleotide hydrolysis and the unwinding activity of G40P are affected in a different manner by different nucleotide cofactors. Nuclease protection studies suggest that G40P protects the 5' tail of a forked molecule, and the duplex region at the junction against exonuclease attack. G40P does not protect the 3' tail of a forked molecule from exonuclease attack. By using electron microscopy we confirm that the ssDNA transverses the centre of the hexameric ring. Our results show that hexameric G40P DNA helicase encircles the 5' tail, interacts with the duplex DNA at the ss-double-stranded DNA junction and excludes the 3' tail of the forked DNA.
Figures



Similar articles
-
Bacillus subtilis bacteriophage SPP1 G40P helicase lacking the n-terminal domain unwinds DNA bidirectionally.J Mol Biol. 2006 Apr 7;357(4):1077-88. doi: 10.1016/j.jmb.2005.12.027. Epub 2005 Dec 27. J Mol Biol. 2006. PMID: 16405907
-
The Bacillus subtilis bacteriophage SPP1 G39P delivers and activates the G40P DNA helicase upon interacting with the G38P-bound replication origin.J Mol Biol. 1999 Apr 23;288(1):71-85. doi: 10.1006/jmbi.1999.2662. J Mol Biol. 1999. PMID: 10329127
-
Bacillus subtilis tau subunit of DNA polymerase III interacts with bacteriophage SPP1 replicative DNA helicase G40P.Nucleic Acids Res. 2002 Dec 1;30(23):5056-64. doi: 10.1093/nar/gkf650. Nucleic Acids Res. 2002. PMID: 12466528 Free PMC article.
-
Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase.Curr Opin Chem Biol. 2011 Oct;15(5):595-605. doi: 10.1016/j.cbpa.2011.08.003. Epub 2011 Aug 22. Curr Opin Chem Biol. 2011. PMID: 21865075 Free PMC article. Review.
-
Non-hexameric DNA helicases and translocases: mechanisms and regulation.Nat Rev Mol Cell Biol. 2008 May;9(5):391-401. doi: 10.1038/nrm2394. Nat Rev Mol Cell Biol. 2008. PMID: 18414490 Review.
Cited by
-
A novel role for RecA under non-stress: promotion of swarming motility in Escherichia coli K-12.BMC Biol. 2007 Mar 28;5:14. doi: 10.1186/1741-7007-5-14. BMC Biol. 2007. PMID: 17391508 Free PMC article.
-
Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery.Eur J Biochem. 2004 May;271(10):1835-48. doi: 10.1111/j.1432-1033.2004.04093.x. Eur J Biochem. 2004. PMID: 15128294 Free PMC article. Review.
-
Bacillus subtilis DNA polymerases, PolC and DnaE, are required for both leading and lagging strand synthesis in SPP1 origin-dependent DNA replication.Nucleic Acids Res. 2017 Aug 21;45(14):8302-8313. doi: 10.1093/nar/gkx493. Nucleic Acids Res. 2017. PMID: 28575448 Free PMC article.
-
Characterization of the Holliday junction resolving enzyme encoded by the Bacillus subtilis bacteriophage SPP1.PLoS One. 2012;7(10):e48440. doi: 10.1371/journal.pone.0048440. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23119018 Free PMC article.
-
Bacillus subtilis RecA interacts with and loads RadA/Sms to unwind recombination intermediates during natural chromosomal transformation.Nucleic Acids Res. 2019 Sep 26;47(17):9198-9215. doi: 10.1093/nar/gkz647. Nucleic Acids Res. 2019. PMID: 31350886 Free PMC article.
References
-
- Pedré X., Weise,F., Chai,S., Lüder,G. and Alonso,J.C. (1994) Characterization of cis and trans acting elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol., 236, 1324–1340. - PubMed
-
- Missich R., Weise,F., Chai,S., Lurz,R., Pedré,X. and Alonso,J.C. (1997) The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol., 270, 50–64. - PubMed
-
- Ayora S., Stasiak,A. and Alonso,J.C. (1999) The Bacillus subtilis bacteriophage SPP1 G39P delivers and activates the G40P DNA helicase upon interacting with the G38P-bound replication origin. J. Mol. Biol., 288, 71–85. - PubMed
-
- Ayora S., Langer,U. and Alonso,J.C. (1998) Bacillus subtilis DnaG primase stabilises the bacteriophage SPP1 G40P helicase-ssDNA complex. FEBS Lett., 439, 59–62. - PubMed
-
- Bárcena M., San Martin,C., Weise,F., Ayora,S., Alonso,J.C. and Carazo,J.M. (1998) Polymorphic quaternary organization of the Bacillus subtilis bacteriophage SPP1 replicative helicase (G40P). J. Mol. Biol., 283, 809–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

