Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation
- PMID: 12024030
- PMCID: PMC133845
- DOI: 10.1128/MCB.22.12.4167-4180.2002
Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation
Abstract
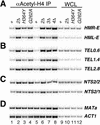
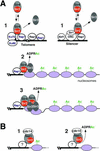
Transcriptional silencing at the budding yeast silent mating type (HM) loci and telomeric DNA regions requires Sir2, a conserved NAD-dependent histone deacetylase, Sir3, Sir4, histones H3 and H4, and several DNA-binding proteins. Silencing at the yeast ribosomal DNA (rDNA) repeats requires a complex containing Sir2, Net1, and Cdc14. Here we show that the native Sir2/Sir4 complex is composed solely of Sir2 and Sir4 and that native Sir3 is not associated with other proteins. We further show that the initial binding of the Sir2/Sir4 complex to DNA sites that nucleate silencing, accompanied by partial Sir2-dependent histone deacetylation, occurs independently of Sir3 and is likely to be the first step in assembly of silent chromatin at the HM loci and telomeres. The enzymatic activity of Sir2 is not required for this initial binding, but is required for the association of silencing proteins with regions distal from nucleation sites. At the rDNA repeats, we show that histone H3 and H4 tails are required for silencing and rDNA-associated H4 is hypoacetylated in a Sir2-dependent manner. However, the binding of Sir2 to rDNA is independent of its histone deacetylase activity. Together, these results support a stepwise model for the assembly of silent chromatin domains in Saccharomyces cerevisiae.
Figures





Similar articles
-
Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase.Nature. 2000 Feb 17;403(6771):795-800. doi: 10.1038/35001622. Nature. 2000. PMID: 10693811
-
A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin.Mol Cell Biol. 2005 Jun;25(11):4514-28. doi: 10.1128/MCB.25.11.4514-4528.2005. Mol Cell Biol. 2005. PMID: 15899856 Free PMC article.
-
A model for step-wise assembly of heterochromatin in yeast.Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review.
-
A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors.Mol Cell Biol. 1999 Apr;19(4):3184-97. doi: 10.1128/MCB.19.4.3184. Mol Cell Biol. 1999. PMID: 10082585 Free PMC article.
-
Enzymatic activities of Sir2 and chromatin silencing.Curr Opin Cell Biol. 2001 Apr;13(2):232-8. doi: 10.1016/s0955-0674(00)00202-7. Curr Opin Cell Biol. 2001. PMID: 11248558 Review.
Cited by
-
Histone Deacetylases with Antagonistic Roles in Saccharomyces cerevisiae Heterochromatin Formation.Genetics. 2016 Sep;204(1):177-90. doi: 10.1534/genetics.116.190835. Epub 2016 Aug 3. Genetics. 2016. PMID: 27489001 Free PMC article.
-
Gene repression in S. cerevisiae-looking beyond Sir-dependent gene silencing.Curr Genet. 2021 Feb;67(1):3-17. doi: 10.1007/s00294-020-01114-7. Epub 2020 Oct 10. Curr Genet. 2021. PMID: 33037902 Review.
-
Unwrap RAP1's Mystery at Kinetoplastid Telomeres.Biomolecules. 2024 Jan 4;14(1):67. doi: 10.3390/biom14010067. Biomolecules. 2024. PMID: 38254667 Free PMC article. Review.
-
Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures.Mol Cell Biol. 2004 Mar;24(5):2118-31. doi: 10.1128/MCB.24.5.2118-2131.2004. Mol Cell Biol. 2004. PMID: 14966290 Free PMC article.
-
Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing.Genetics. 2004 Sep;168(1):547-51. doi: 10.1534/genetics.104.028803. Genetics. 2004. PMID: 15454564 Free PMC article.
References
-
- Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. - PubMed
-
- Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
