Induction of the bovine papillomavirus origin "onion skin"-type DNA replication at high E1 protein concentrations in vivo
- PMID: 11992014
- PMCID: PMC137012
- DOI: 10.1128/jvi.76.11.5835-5845.2002
Induction of the bovine papillomavirus origin "onion skin"-type DNA replication at high E1 protein concentrations in vivo
Abstract
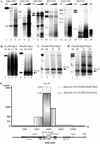
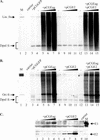
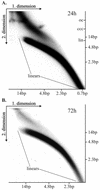
We have studied the replication of plasmids composed of bovine papillomavirus type 1 (BPV1) origin of replication and expression cartridges for viral proteins E1 and E2 in hamster and mouse cells. We found that the replication mode changed dramatically at different expression levels of the E1 protein. At high levels of the E1 protein, overreplication of the origin region of the plasmid was observed. Analysis of the replication products by one-dimensional and two-dimensional gel electrophoresis suggested that initially "onion skin"-type replication intermediates were generated, presumably resulting from initiation of the new replication forks before the leading fork completed the synthesis of the DNA on the episomal plasmid. These replication intermediates served as templates for generation of a heterogeneous set of origin region-containing linear fragments by displacement synthesis at the partially replicated plasmid. Additionally, the linear fragments may have been generated by DNA break-up of the onion skin-type intermediates. Analysis of replication products indicated that generated linear fragments recombined and formed concatemers or circular molecules, which presumably were able to replicate in an E1- and E2-dependent fashion. At moderate and low levels of E1, generated by transcription of the E1 open reading frame using weaker promoters, DNA replication was initiated at much lower levels, which allowed elongation of the replication fork starting from the origin to be more balanced and resulted in the generation of full-sized replication products.
Figures



Similar articles
-
E1 protein of human papillomavirus type 1a is sufficient for initiation of viral DNA replication.Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9597-601. doi: 10.1073/pnas.91.20.9597. Proc Natl Acad Sci U S A. 1994. PMID: 7937813 Free PMC article.
-
Bovine papillomavirus type 1 DNA replication: the transcriptional activator E2 acts in vitro as a specificity factor.J Virol. 1997 Sep;71(9):6805-15. doi: 10.1128/JVI.71.9.6805-6815.1997. J Virol. 1997. PMID: 9261405 Free PMC article.
-
The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase.Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5086-90. doi: 10.1073/pnas.90.11.5086. Proc Natl Acad Sci U S A. 1993. PMID: 8389467 Free PMC article.
-
E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication.Virology. 2002 Feb 1;293(1):10-4. doi: 10.1006/viro.2001.1305. Virology. 2002. PMID: 11853393
-
The papillomavirus E2 protein: a factor with many talents.Trends Biochem Sci. 1991 Nov;16(11):440-4. doi: 10.1016/0968-0004(91)90172-r. Trends Biochem Sci. 1991. PMID: 1663669 Review.
Cited by
-
Replication and partitioning of papillomavirus genomes.Adv Virus Res. 2008;72:155-205. doi: 10.1016/S0065-3527(08)00404-1. Adv Virus Res. 2008. PMID: 19081491 Free PMC article. Review.
-
Gene Therapy: The Potential Applicability of Gene Transfer Technology to the Human Germline.Int J Med Sci. 2004;1(2):76-91. doi: 10.7150/ijms.1.76. Epub 2004 Jun 1. Int J Med Sci. 2004. PMID: 15912200 Free PMC article.
-
The Causes and Consequences of DNA Damage and Chromosomal Instability Induced by Human Papillomavirus.Cancers (Basel). 2024 Apr 25;16(9):1662. doi: 10.3390/cancers16091662. Cancers (Basel). 2024. PMID: 38730612 Free PMC article. Review.
-
Oncogenic activities of human papillomaviruses.Virus Res. 2009 Aug;143(2):195-208. doi: 10.1016/j.virusres.2009.06.008. Epub 2009 Jun 18. Virus Res. 2009. PMID: 19540281 Free PMC article. Review.
-
Regulation of human papillomavirus gene expression by splicing and polyadenylation.Nat Rev Microbiol. 2013 Apr;11(4):239-51. doi: 10.1038/nrmicro2984. Epub 2013 Mar 11. Nat Rev Microbiol. 2013. PMID: 23474685 Review.
References
-
- Amin, A. A., S. Titolo, A. Pelletier, D. Fink, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the HPV11 E1 protein required for DNA replication in vitro. Virology 272:137-150. - PubMed
-
- Belyavskyi, M., M. Westerman, L. DiMichele, and V. Wilson. 1996. Perturbation of the host cell cycle and DNA replication by the bovine papillomavirus replication protein E1. Virology 219:206-219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

