Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase
- PMID: 11971977
- PMCID: PMC133788
- DOI: 10.1128/MCB.22.10.3460-3473.2002
Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase
Abstract
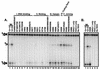
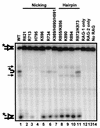
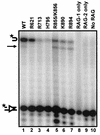
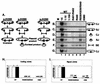
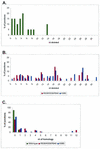
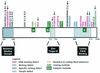
Although both RAG-1 and RAG-2 are required for all steps of V(D)J recombination, little is known about the specific contribution of either protein to these steps. RAG-1 contains three acidic active-site amino acids that are thought to coordinate catalytic metal ions. To search for additional catalytic amino acids and to better define the functional anatomy of RAG-1, we mutated all 86 conserved basic amino acids to alanine and evaluated the mutant proteins for DNA binding, nicking, hairpin formation, and joining. We found several amino acids outside of the canonical nonamer-binding domain that are critical for DNA binding, several step arrest mutants with defects in nicking or hairpin formation, and four RAG-1 mutants defective specifically for joining. Analysis of coding joints formed by some of these mutants revealed excessive deletions, frequent use of short sequence homologies, and unusually long palindromic junctional inserts, known as P nucleotides, that result from aberrant hairpin opening. These features characterize junctions found in scid mice, which are deficient for the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), suggesting that the RAG proteins and DNA-PKcs perform overlapping functions in coding joint formation. Interestingly, the amino acids that are altered in 12 of our mutants are also mutated in human inherited immunodeficiency syndromes. Our analysis of these mutants provides insights into the molecular mechanisms underlying these disorders.
Figures

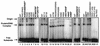

Similar articles
-
The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination.Mol Cell Biol. 2001 Jan;21(2):449-58. doi: 10.1128/MCB.21.2.449-458.2001. Mol Cell Biol. 2001. PMID: 11134333 Free PMC article.
-
Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination.Genes Dev. 1999 Dec 1;13(23):3059-69. doi: 10.1101/gad.13.23.3059. Genes Dev. 1999. PMID: 10601032 Free PMC article.
-
Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro.Mol Cell. 2001 Jan;7(1):65-75. doi: 10.1016/s1097-2765(01)00155-1. Mol Cell. 2001. PMID: 11172712
-
The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu Rev Immunol. 2000;18:495-527. doi: 10.1146/annurev.immunol.18.1.495. Annu Rev Immunol. 2000. PMID: 10837067 Review.
-
RAG1 and RAG2 in V(D)J recombination and transposition.Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review.
Cited by
-
Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways.Nucleic Acids Res. 2010 May;38(9):2944-54. doi: 10.1093/nar/gkp1252. Epub 2010 Feb 4. Nucleic Acids Res. 2010. PMID: 20139091 Free PMC article.
-
Molecular analysis of T-B-NK+ severe combined immunodeficiency and Omenn syndrome cases in Saudi Arabia.BMC Med Genet. 2009 Nov 13;10:116. doi: 10.1186/1471-2350-10-116. BMC Med Genet. 2009. PMID: 19912631 Free PMC article.
-
Full-length RAG1 promotes contact with coding and intersignal sequences in RAG protein complexes bound to recombination signals paired in cis.Nucleic Acids Res. 2009 Apr;37(7):2211-26. doi: 10.1093/nar/gkp047. Epub 2009 Feb 20. Nucleic Acids Res. 2009. PMID: 19233873 Free PMC article.
-
Crystal structure of the V(D)J recombinase RAG1-RAG2.Nature. 2015 Feb 26;518(7540):507-11. doi: 10.1038/nature14174. Epub 2015 Feb 18. Nature. 2015. PMID: 25707801 Free PMC article.
-
Understanding how the V(D)J recombinase catalyzes transesterification: distinctions between DNA cleavage and transposition.Nucleic Acids Res. 2008 May;36(9):2864-73. doi: 10.1093/nar/gkn128. Epub 2008 Mar 29. Nucleic Acids Res. 2008. PMID: 18375979 Free PMC article.
References
-
- Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. - PubMed
-
- Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. - PubMed
-
- Bogue, M. A., C. Wang, C. Zhu, and D. B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7:37-47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
