The small viral membrane-associated protein P32 is involved in bacteriophage PRD1 DNA entry
- PMID: 11967303
- PMCID: PMC136160
- DOI: 10.1128/jvi.76.10.4866-4872.2002
The small viral membrane-associated protein P32 is involved in bacteriophage PRD1 DNA entry
Abstract
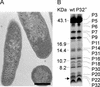
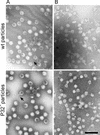
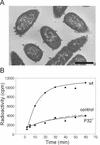
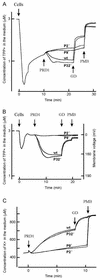
The lipid-containing bacteriophage PRD1 infects a variety of gram-negative cells by injecting its linear double-stranded DNA genome into the host cell cytoplasm, while the protein capsid is left outside. The virus membrane and several structural proteins are involved in phage DNA entry. In this work we identified a new infectivity protein of PRD1. Disruption of gene XXXII resulted in a mutant phenotype defective in phage reproduction. The absence of the protein P32 did not compromise the particle assembly but led to a defect in phage DNA injection. In P32-deficient particles the phage membrane is unable to undergo a structural transformation from a spherical to a tubular form. Since P32(-) particles are able to increase the permeability of the host cell envelope to a degree comparable to that found with wild-type particles, we suggest that the tail-tube formation is needed to eject the DNA from the phage particle rather than to reach the host cell interior.
Figures

Similar articles
-
Integral membrane protein P16 of bacteriophage PRD1 stabilizes the adsorption vertex structure.J Virol. 2004 Sep;78(18):9790-7. doi: 10.1128/JVI.78.18.9790-9797.2004. J Virol. 2004. PMID: 15331712 Free PMC article.
-
Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane.Mol Microbiol. 2002 Dec;46(5):1199-209. doi: 10.1046/j.1365-2958.2002.03250.x. Mol Microbiol. 2002. PMID: 12453208
-
The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane.J Bacteriol. 2002 Jan;184(1):104-10. doi: 10.1128/JB.184.1.104-110.2002. J Bacteriol. 2002. PMID: 11741849 Free PMC article.
-
Lipid-containing viruses: bacteriophage PRD1 assembly.Adv Exp Med Biol. 2012;726:365-77. doi: 10.1007/978-1-4614-0980-9_16. Adv Exp Med Biol. 2012. PMID: 22297522 Review.
-
Membrane-Containing Icosahedral Bacteriophage PRD1: The Dawn of Viral Lineages.Adv Exp Med Biol. 2019;1215:85-109. doi: 10.1007/978-3-030-14741-9_5. Adv Exp Med Biol. 2019. PMID: 31317497 Review.
Cited by
-
Archaeal viruses at the cell envelope: entry and egress.Front Microbiol. 2015 Jun 5;6:552. doi: 10.3389/fmicb.2015.00552. eCollection 2015. Front Microbiol. 2015. PMID: 26097469 Free PMC article. Review.
-
The unique vertex of bacterial virus PRD1 is connected to the viral internal membrane.J Virol. 2003 Jun;77(11):6314-21. doi: 10.1128/jvi.77.11.6314-6321.2003. J Virol. 2003. PMID: 12743288 Free PMC article.
-
Mechanism of membranous tunnelling nanotube formation in viral genome delivery.PLoS Biol. 2013 Sep;11(9):e1001667. doi: 10.1371/journal.pbio.1001667. Epub 2013 Sep 24. PLoS Biol. 2013. PMID: 24086111 Free PMC article.
-
The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex.J Virol. 2003 Jul;77(14):7863-71. doi: 10.1128/jvi.77.14.7863-7871.2003. J Virol. 2003. PMID: 12829826 Free PMC article.
-
Integral membrane protein P16 of bacteriophage PRD1 stabilizes the adsorption vertex structure.J Virol. 2004 Sep;78(18):9790-7. doi: 10.1128/JVI.78.18.9790-9797.2004. J Virol. 2004. PMID: 15331712 Free PMC article.
References
-
- Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. - PubMed
-
- Bamford, D. H., and L. Mindich. 1982. Electron microscopy of cells infected with nonsense mutants of bacteriophage Φ6. Virology 710:222-228. - PubMed
-
- Bamford, D. H., L. Rouhiainen, K. Takkinen, and H. Söderlund. 1981. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J. Gen. Virol. 57:365-373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

