Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence
- PMID: 11891244
- PMCID: PMC152201
- DOI: 10.1104/pp.010843
Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence
Abstract
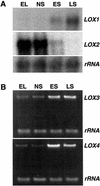
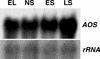
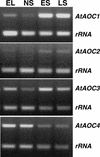
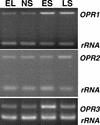
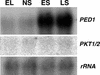
In this work, the role of jasmonic acid (JA) in leaf senescence is examined. Exogenous application of JA caused premature senescence in attached and detached leaves in wild-type Arabidopsis but failed to induce precocious senescence of JA-insensitive mutant coi1 plants, suggesting that the JA-signaling pathway is required for JA to promote leaf senescence. JA levels in senescing leaves are 4-fold higher than in non-senescing ones. Concurrent with the increase in JA level in senescing leaves, genes encoding the enzymes that catalyze most of the reactions of the JA biosynthetic pathway are differentially activated during leaf senescence in Arabidopsis, except for allene oxide synthase, which is constitutively and highly expressed throughout leaf development. Arabidopsis lipoxygenase 1 (cytoplasmic) expression is greatly increased but lipoxygenase 2 (plastidial) expression is sharply reduced during leaf senescence. Similarly, AOC1 (allene oxide cyclase 1), AOC2, and AOC3 are all up-regulated, whereas AOC4 is down-regulated with the progression of leaf senescence. The transcript levels of 12-oxo-PDA reductase 1 and 12-oxo-PDA reductase 3 also increase in senescing leaves, as does PED1 (encoding a 3-keto-acyl-thiolase for beta-oxidation). This represents the first report, to our knowledge, of an increase in JA levels and expression of oxylipin genes during leaf senescence, and indicates that JA may play a role in the senescence program.
Figures


Similar articles
-
Identical promoter elements are involved in regulation of the OPR1 gene by senescence and jasmonic acid in Arabidopsis.Plant Mol Biol. 2001 Nov;47(5):595-605. doi: 10.1023/a:1012211011538. Plant Mol Biol. 2001. PMID: 11725945
-
ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: tissue- and organ-specific promoter activities and in vivo heteromerization.J Exp Bot. 2012 Oct;63(17):6125-38. doi: 10.1093/jxb/ers261. Epub 2012 Oct 1. J Exp Bot. 2012. PMID: 23028017 Free PMC article.
-
Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana.Plant Mol Biol. 2003 Apr;51(6):895-911. doi: 10.1023/a:1023049319723. Plant Mol Biol. 2003. PMID: 12777050
-
Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones.J Exp Bot. 2017 Mar 1;68(6):1361-1369. doi: 10.1093/jxb/erx004. J Exp Bot. 2017. PMID: 28201612 Review.
-
Rice octadecanoid pathway.Biochem Biophys Res Commun. 2004 Apr 23;317(1):1-15. doi: 10.1016/j.bbrc.2004.03.020. Biochem Biophys Res Commun. 2004. PMID: 15047141 Review.
Cited by
-
Functional Analysis of Jasmonates in Rice through Mutant Approaches.Plants (Basel). 2016 Mar 18;5(1):15. doi: 10.3390/plants5010015. Plants (Basel). 2016. PMID: 27135235 Free PMC article. Review.
-
A Clade-Specific Arabidopsis Gene Connects Primary Metabolism and Senescence.Front Plant Sci. 2016 Jul 12;7:983. doi: 10.3389/fpls.2016.00983. eCollection 2016. Front Plant Sci. 2016. PMID: 27462324 Free PMC article.
-
Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence.Photosynth Res. 2013 Nov;117(1-3):221-34. doi: 10.1007/s11120-013-9862-x. Epub 2013 Jun 15. Photosynth Res. 2013. PMID: 23771643 Review.
-
The oxylipin pathway in Arabidopsis.Arabidopsis Book. 2002;1:e0012. doi: 10.1199/tab.0012. Epub 2002 Aug 12. Arabidopsis Book. 2002. PMID: 22303193 Free PMC article.
-
Molecular, in silico and expression studies on lipoxygenases (LOXs) in potato (Solanum tuberosum L.).3 Biotech. 2023 Dec;13(12):419. doi: 10.1007/s13205-023-03839-x. Epub 2023 Nov 28. 3 Biotech. 2023. PMID: 38037658 Free PMC article.
References
-
- Albrecht T, Kehlen A, Stahl K, Knöfel H-D, Sembdner G, Weiler EW. Quantification of rapid, transient increases in jasmonic acid in wounded plants using a monoclonal antibody. Planta. 1993;191:86–94.
-
- Biesgen C, Weiler EW. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta. 1999;208:155–165. - PubMed
-
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

