Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A
- PMID: 11884592
- PMCID: PMC133689
- DOI: 10.1128/MCB.22.7.2037-2046.2002
Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A
Abstract
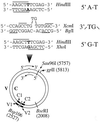
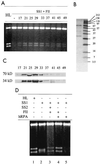
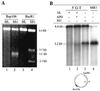
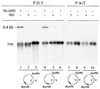
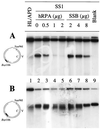
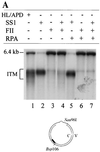
DNA mismatch repair (MMR) is a critical genome-stabilization system. However, the molecular mechanism of MMR in human cells remains obscure because many of the components have not yet been identified. Using a functional in vitro reconstitution system, this study identified three HeLa cell fractions essential for in vitro MMR. These fractions divide human MMR into two distinct stages: mismatch-provoked excision and repair synthesis. In vitro dissection of the MMR reaction and crucial intermediates elucidated biochemical functions of individual fractions in human MMR and identified hitherto unknown functions of human replication protein A (hRPA) in MMR. Thus, one fraction carries out nick-directed and mismatch-dependent excision; the second carries out DNA repair synthesis and DNA ligation; and the third provides hRPA, which plays multiple roles in human MMR by protecting the template DNA strand from degradation, enhancing repair excision, and facilitating repair synthesis. It is anticipated that further analysis of these fractions will identify additional MMR components and enable the complete reconstitution of the human MMR pathway with purified proteins.
Figures




Similar articles
-
Construction and characterization of mismatch-containing circular DNA molecules competent for assessment of nick-directed human mismatch repair in vitro.Nucleic Acids Res. 2002 Feb 1;30(3):E14. doi: 10.1093/nar/30.3.e14. Nucleic Acids Res. 2002. PMID: 11809902 Free PMC article.
-
Regulation of replication protein A functions in DNA mismatch repair by phosphorylation.J Biol Chem. 2006 Aug 4;281(31):21607-21616. doi: 10.1074/jbc.M603504200. Epub 2006 May 26. J Biol Chem. 2006. PMID: 16731533
-
Influence of oxidized purine processing on strand directionality of mismatch repair.J Biol Chem. 2015 Apr 17;290(16):9986-99. doi: 10.1074/jbc.M114.629907. Epub 2015 Feb 18. J Biol Chem. 2015. PMID: 25694431 Free PMC article.
-
Functional interactions and signaling properties of mammalian DNA mismatch repair proteins.Cell Death Differ. 2001 Nov;8(11):1076-92. doi: 10.1038/sj.cdd.4400948. Cell Death Differ. 2001. PMID: 11687886 Review.
-
Mismatch repair proteins: key regulators of genetic recombination.Cytogenet Genome Res. 2004;107(3-4):146-59. doi: 10.1159/000080593. Cytogenet Genome Res. 2004. PMID: 15467360 Review.
Cited by
-
DNA mismatch repair in cancer immunotherapy.NAR Cancer. 2023 Jun 13;5(3):zcad031. doi: 10.1093/narcan/zcad031. eCollection 2023 Sep. NAR Cancer. 2023. PMID: 37325548 Free PMC article.
-
The mechanism of mismatch repair and the functional analysis of mismatch repair defects in Lynch syndrome.Fam Cancer. 2013 Jun;12(2):159-68. doi: 10.1007/s10689-013-9635-x. Fam Cancer. 2013. PMID: 23572416 Free PMC article. Review.
-
Unboxing the molecular modalities of mutagens in cancer.Environ Sci Pollut Res Int. 2022 Sep;29(41):62111-62159. doi: 10.1007/s11356-021-16726-w. Epub 2021 Oct 5. Environ Sci Pollut Res Int. 2022. PMID: 34611806 Free PMC article. Review.
-
Distinct structural alterations in proliferating cell nuclear antigen block DNA mismatch repair.Biochemistry. 2013 Aug 20;52(33):5611-9. doi: 10.1021/bi400378e. Epub 2013 Aug 2. Biochemistry. 2013. PMID: 23869605 Free PMC article.
-
The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)(n)/(CTG)(n) hairpins.J Biol Chem. 2012 Aug 31;287(36):30151-6. doi: 10.1074/jbc.M112.389791. Epub 2012 Jul 11. J Biol Chem. 2012. PMID: 22787159 Free PMC article.
References
-
- Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. N. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859-868. - PubMed
-
- Bowers, J., P. T. Tran, A. Joshi, R. M. Liskay, and E. Alani. 2001. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol. 306:957-968. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Brill, S. J., and B. Stillman. 1989. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature 342:92-95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
