The 3' end formation in small RNAs
- PMID: 11868988
- PMCID: PMC5977532
The 3' end formation in small RNAs
Abstract
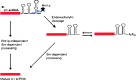
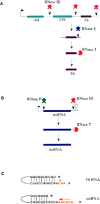
Small RNAs are a major class of RNAs along with transfer RNAs, ribosomal RNAs, and messenger RNAs. They vary in size from less than 100 nucleotides to several thousand nucleotides and have been identified and characterized both in prokaryotes and eukaryotes. Small RNAs participate in a variety of cellular functions including regulating RNA synthesis, RNA processing, guiding modifications in RNA, and in transport of proteins. Small RNAs are generated by a series of posttranscriptional processing steps following transcription. While RNA 5' end structure, 5' cap formation, and RNA processing mechanisms have been fairly well characterized, the 3' end processing is poorly understood. Recent data point to an emerging theme in small RNAs metabolism in which the 3' end processing is mediated by the exosome, a large multienzyme complex. In addition to removal of nucleotides by the exosome, there is simultaneous rebuilding of the 3' end of some small RNA by adenylation and/or uridylation. This review presents a picture of both degradative and rebuilding reactions operative on the 3' end of some small RNA molecules in prokaryotes and eukaryotes.
Figures


Similar articles
-
Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs.Mol Cell Biol. 2000 Jan;20(2):441-52. doi: 10.1128/MCB.20.2.441-452.2000. Mol Cell Biol. 2000. PMID: 10611222 Free PMC article.
-
Disruption of the 5' stem-loop of yeast U6 RNA induces trimethylguanosine capping of this RNA polymerase III transcript in vivo.RNA. 2000 Dec;6(12):1859-69. doi: 10.1017/s1355838200991325. RNA. 2000. PMID: 11142384 Free PMC article.
-
From transcription to mRNA: PAF provides a new path.Mol Cell. 2005 Oct 28;20(2):167-8. doi: 10.1016/j.molcel.2005.10.004. Mol Cell. 2005. PMID: 16246718 Review.
-
Origins and activities of the eukaryotic exosome.J Cell Sci. 2009 May 15;122(Pt 10):1487-94. doi: 10.1242/jcs.047399. J Cell Sci. 2009. PMID: 19420235 Review.
-
3' processing of eukaryotic precursor tRNAs.Wiley Interdiscip Rev RNA. 2011 May-Jun;2(3):362-75. doi: 10.1002/wrna.64. Wiley Interdiscip Rev RNA. 2011. PMID: 21572561 Free PMC article. Review.
Cited by
-
3'-End polishing of the kinetoplastid spliced leader RNA is performed by SNIP, a 3'-->5' exonuclease with a Motley assortment of small RNA substrates.Mol Cell Biol. 2004 Dec;24(23):10390-6. doi: 10.1128/MCB.24.23.10390-10396.2004. Mol Cell Biol. 2004. PMID: 15542846 Free PMC article.
-
trans and cis splicing in trypanosomatids: mechanism, factors, and regulation.Eukaryot Cell. 2003 Oct;2(5):830-40. doi: 10.1128/EC.2.5.830-840.2003. Eukaryot Cell. 2003. PMID: 14555465 Free PMC article. Review. No abstract available.
-
Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome.RNA. 2006 Oct;12(10):1810-6. doi: 10.1261/rna.144606. Epub 2006 Aug 15. RNA. 2006. PMID: 16912217 Free PMC article.
-
Tailer: a pipeline for sequencing-based analysis of nonpolyadenylated RNA 3' end processing.RNA. 2022 May;28(5):645-656. doi: 10.1261/rna.079071.121. Epub 2022 Feb 18. RNA. 2022. PMID: 35181644 Free PMC article.
-
Altered rRNA processing disrupts nuclear RNA homeostasis via competition for the poly(A)-binding protein Nab2.Nucleic Acids Res. 2020 Nov 18;48(20):11675-11694. doi: 10.1093/nar/gkaa964. Nucleic Acids Res. 2020. PMID: 33137177 Free PMC article.
References
-
- Balakin A. G.; Smith L.; Fournier M. J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86:823–834; 1996. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
