Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors
- PMID: 11861919
- PMCID: PMC101241
- DOI: 10.1093/nar/30.5.1255
Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors
Abstract
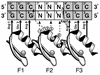
We can determine the effects of many possible sequence variations in transcription factor binding sites using microarray binding experiments. Analysis of wild-type and mutant Zif268 (Egr1) zinc fingers bound to microarrays containing all possible central 3 bp triplet binding sites indicates that the nucleotides of transcription factor binding sites cannot be treated independently. This indicates that the current practice of characterizing transcription factor binding sites by mutating individual positions of binding sites one base pair at a time does not provide a true picture of the sequence specificity. Similarly, current bioinformatic practices using either just a consensus sequence, or even mononucleotide frequency weight matrices to provide more complete descriptions of transcription factor binding sites, are not accurate in depicting the true binding site specificities, since these methods rely upon the assumption that the nucleotides of binding sites exert independent effects on binding affinity. Our results stress the importance of complete reference tables of all possible binding sites for comparing protein binding preferences for various DNA sequences. We also show results suggesting that microarray binding data using particular subsets of all possible binding sites can be used to extrapolate the relative binding affinities of all possible full-length binding sites, given a known binding site for use as a starting sequence for site preference refinement.
Figures
Similar articles
-
Exploring the DNA-binding specificities of zinc fingers with DNA microarrays.Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7158-63. doi: 10.1073/pnas.111163698. Epub 2001 Jun 12. Proc Natl Acad Sci U S A. 2001. PMID: 11404456 Free PMC article.
-
Mechanism of DNA binding by the ADR1 zinc finger transcription factor as determined by SPR.J Mol Biol. 2003 Jun 20;329(5):931-9. doi: 10.1016/s0022-2836(03)00550-3. J Mol Biol. 2003. PMID: 12798683
-
High affinity binding sites for the Wilms' tumour suppressor protein WT1.Nucleic Acids Res. 1995 Jan 25;23(2):277-84. doi: 10.1093/nar/23.2.277. Nucleic Acids Res. 1995. PMID: 7862533 Free PMC article.
-
Accuracy and reproducibility of protein-DNA microarray technology.Adv Biochem Eng Biotechnol. 2007;104:87-110. doi: 10.1007/10_2006_035. Adv Biochem Eng Biotechnol. 2007. PMID: 17290820 Review.
-
[Double-stranded DNA microarray: principal, techniques and applications].Yi Chuan. 2013 Mar;35(3):287-306. doi: 10.3724/sp.j.1005.2013.00287. Yi Chuan. 2013. PMID: 23575535 Review. Chinese.
Cited by
-
Effect of positional dependence and alignment strategy on modeling transcription factor binding sites.BMC Res Notes. 2012 Jul 2;5:340. doi: 10.1186/1756-0500-5-340. BMC Res Notes. 2012. PMID: 22748199 Free PMC article.
-
PairMotif: A new pattern-driven algorithm for planted (l, d) DNA motif search.PLoS One. 2012;7(10):e48442. doi: 10.1371/journal.pone.0048442. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23119020 Free PMC article.
-
Protein binding microarrays for the characterization of DNA-protein interactions.Adv Biochem Eng Biotechnol. 2007;104:65-85. doi: 10.1007/10_025. Adv Biochem Eng Biotechnol. 2007. PMID: 17290819 Free PMC article. Review.
-
Metamotifs--a generative model for building families of nucleotide position weight matrices.BMC Bioinformatics. 2010 Jun 25;11:348. doi: 10.1186/1471-2105-11-348. BMC Bioinformatics. 2010. PMID: 20579334 Free PMC article.
-
A combinatorial optimization approach for diverse motif finding applications.Algorithms Mol Biol. 2006 Aug 17;1:13. doi: 10.1186/1748-7188-1-13. Algorithms Mol Biol. 2006. PMID: 16916460 Free PMC article.
References
-
- Staden R. (1988) Methods to define and locate patterns of motifs in sequences. Comput. Appl. Biosci., 4, 53–60. - PubMed
-
- Zhang M.Q. and Marr,T.G. (1993) A weight array method for splicing signal analysis. Comput. Appl. Biosci., 9, 499–509. - PubMed
-
- Ponomarenko M.P., Ponomarenko,J.V., Frolov,A.S., Podkolodnaya,O.A., Vorobyev,D.G., Kolchanov,N.A. and Overton,G.C. (1999) Oligonucleotide frequency matrices addressed to recognizing functional DNA sites. Bioinformatics, 15, 631–643. - PubMed
-
- Wingender E., Karas,H. and Knuppel,R. (1997) TRANSFAC database as a bridge between sequence data libraries and biological function. Pac. Symp. Biocomput., 477–485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

