Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro)
- PMID: 11836431
- PMCID: PMC135932
- DOI: 10.1128/jvi.76.5.2529-2542.2002
Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro)
Abstract
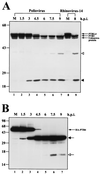
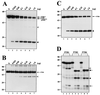
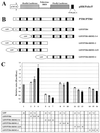
The translation of polioviral mRNA occurs through an internal ribosomal entry site (IRES). Several RNA-binding proteins, such as polypyrimidine tract-binding protein (PTB) and poly(rC)-binding protein (PCBP), are required for the poliovirus IRES-dependent translation. Here we report that a poliovirus protein, 3C(pro) (and/or 3CD(pro)), cleaves PTB isoforms (PTB1, PTB2, and PTB4). Three 3C(pro) target sites (one major target site and two minor target sites) exist in PTBs. PTB fragments generated by poliovirus infection are redistributed to the cytoplasm from the nucleus, where most of the intact PTBs are localized. Moreover, these PTB fragments inhibit polioviral IRES-dependent translation in a cell-based assay system. We speculate that the proteolytic cleavage of PTBs may contribute to the molecular switching from translation to replication of polioviral RNA.
Figures





Similar articles
-
Polypyrimidine tract-binding proteins are cleaved by caspase-3 during apoptosis.J Biol Chem. 2002 Jul 26;277(30):27200-9. doi: 10.1074/jbc.M203887200. Epub 2002 May 9. J Biol Chem. 2002. PMID: 12004072
-
Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome.J Virol. 2000 Mar;74(5):2219-26. doi: 10.1128/jvi.74.5.2219-2226.2000. J Virol. 2000. PMID: 10666252 Free PMC article.
-
Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site.J Virol. 2007 Sep;81(18):10017-28. doi: 10.1128/JVI.00516-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609276 Free PMC article.
-
Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease.J Virol. 2002 Mar;76(5):2062-74. doi: 10.1128/jvi.76.5.2062-2074.2002. J Virol. 2002. PMID: 11836384 Free PMC article.
-
A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein.Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7642-6. doi: 10.1073/pnas.90.16.7642. Proc Natl Acad Sci U S A. 1993. PMID: 8395052 Free PMC article.
Cited by
-
Translational control by viral proteinases.Virus Res. 2006 Jul;119(1):76-88. doi: 10.1016/j.virusres.2005.10.016. Epub 2005 Nov 21. Virus Res. 2006. PMID: 16303201 Free PMC article. Review.
-
Seneca Valley Virus 3C pro Cleaves Heterogeneous Nuclear Ribonucleoprotein K to Facilitate Viral Replication.Front Microbiol. 2022 Jul 6;13:945443. doi: 10.3389/fmicb.2022.945443. eCollection 2022. Front Microbiol. 2022. PMID: 35875542 Free PMC article.
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
-
Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification.J Virol. 2013 Aug;87(15):8697-712. doi: 10.1128/JVI.00950-13. Epub 2013 Jun 5. J Virol. 2013. PMID: 23740976 Free PMC article.
-
Enterovirus Control of Translation and RNA Granule Stress Responses.Viruses. 2016 Mar 30;8(4):93. doi: 10.3390/v8040093. Viruses. 2016. PMID: 27043612 Free PMC article. Review.
References
-
- Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. - PubMed
-
- Andino, R., N. Böddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. - PubMed
-
- Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5" end of poliovirus RNA. Cell 63:369-380. - PubMed
-
- Baltimore, D. 1968. Structure of the poliovirus replicative intermediate RNA. J. Mol. Biol. 32:359-368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

