Dynamic association of transcriptional activation domains and regulatory regions in Saccharomyces cerevisiae heat shock factor
- PMID: 11818569
- PMCID: PMC122167
- DOI: 10.1073/pnas.032681299
Dynamic association of transcriptional activation domains and regulatory regions in Saccharomyces cerevisiae heat shock factor
Abstract
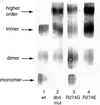
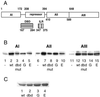
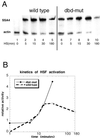
In Saccharomyces cerevisiae, the heat shock transcription factor (HSF) is thought to be a homotypic trimer that is bound to the promoters of heat shock protein (HSP) genes at both normal and heat shock temperatures. Exposure to heat shock greatly and rapidly induces HSF transcriptional activity without further increasing DNA-binding affinity. It is believed that HSF is under negative regulation at normal growth temperatures, but the detailed mechanism by which HSF is activated is still not clear. We report the analysis of mutations in a conserved arginine (residue 274) at the C-terminal end of the DNA-binding domain (DBD). Two mutations significantly increase both basal activity of HSF at normal temperatures and induced activity on heat shock. We demonstrate by coimmunoprecipitation experiments that the mutations reduce the association between the DNA-binding domain/oligomerization domain and the transcription activation domains. Our studies suggest that the DNA-binding domain of HSF can interact with activation domains directly, and this interaction is important for the repression of HSF activity under normal growth conditions. Destabilizing this interaction by heat or by mutations results in HSF transcriptional activation. We propose that Arg-274 is critical for intramolecular repression of HSF activity in normally growing cells.
Figures


Similar articles
-
The C-terminal hydrophobic repeat of Schizosaccharomyces pombe heat shock factor is not required for heat-induced DNA-binding.Yeast. 1998 Jun 15;14(8):733-46. doi: 10.1002/(SICI)1097-0061(19980615)14:8<733::AID-YEA270>3.0.CO;2-8. Yeast. 1998. PMID: 9675818
-
The wing in yeast heat shock transcription factor (HSF) DNA-binding domain is required for full activity.Nucleic Acids Res. 2001 Apr 15;29(8):1715-23. doi: 10.1093/nar/29.8.1715. Nucleic Acids Res. 2001. PMID: 11292844 Free PMC article.
-
Temperature-dependent regulation of a heterologous transcriptional activation domain fused to yeast heat shock transcription factor.Mol Cell Biol. 1992 Mar;12(3):1021-30. doi: 10.1128/mcb.12.3.1021-1030.1992. Mol Cell Biol. 1992. PMID: 1545786 Free PMC article.
-
Heat shock transcription factors: structure and regulation.Annu Rev Cell Dev Biol. 1995;11:441-69. doi: 10.1146/annurev.cb.11.110195.002301. Annu Rev Cell Dev Biol. 1995. PMID: 8689565 Review.
-
HSF transcription factor family, heat shock response, and protein intrinsic disorder.Curr Protein Pept Sci. 2012 Feb;13(1):86-103. doi: 10.2174/138920312799277956. Curr Protein Pept Sci. 2012. PMID: 22044151 Review.
Cited by
-
A functional module of yeast mediator that governs the dynamic range of heat-shock gene expression.Genetics. 2006 Apr;172(4):2169-84. doi: 10.1534/genetics.105.052738. Epub 2006 Feb 1. Genetics. 2006. PMID: 16452140 Free PMC article.
-
Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice.Plant Cell. 2009 Dec;21(12):4031-43. doi: 10.1105/tpc.109.066902. Epub 2009 Dec 22. Plant Cell. 2009. PMID: 20028842 Free PMC article.
-
De novo appearance and "strain" formation of yeast prion [PSI+] are regulated by the heat-shock transcription factor.Genetics. 2006 May;173(1):35-47. doi: 10.1534/genetics.105.054221. Epub 2006 Feb 1. Genetics. 2006. PMID: 16452152 Free PMC article.
-
The conserved PBAF nucleosome-remodeling complex mediates the response to stress in Caenorhabditis elegans.Mol Cell Biol. 2014 Mar;34(6):1121-35. doi: 10.1128/MCB.01502-13. Epub 2014 Jan 13. Mol Cell Biol. 2014. PMID: 24421384 Free PMC article.
-
Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element.Mol Cell Biol. 2004 May;24(9):3648-59. doi: 10.1128/MCB.24.9.3648-3659.2004. Mol Cell Biol. 2004. PMID: 15082761 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

