Involvement of rhp23, a Schizosaccharomyces pombe homolog of the human HHR23A and Saccharomyces cerevisiae RAD23 nucleotide excision repair genes, in cell cycle control and protein ubiquitination
- PMID: 11788722
- PMCID: PMC99819
- DOI: 10.1093/nar/30.2.581
Involvement of rhp23, a Schizosaccharomyces pombe homolog of the human HHR23A and Saccharomyces cerevisiae RAD23 nucleotide excision repair genes, in cell cycle control and protein ubiquitination
Abstract
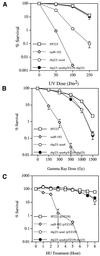
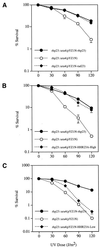
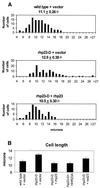
A functional homolog (rhp23) of human HHR23A and Saccharomyces cerevisiae RAD23 was cloned from the fission yeast Schizosaccharomyces pombe and characterized. Consistent with the role of Rad23 homologs in nucleotide excision repair, rhp23 mutant cells are moderately sensitive to UV light but demonstrate wild-type resistance to gamma-rays and hydroxyurea. Expression of the rhp23, RAD23 or HHR23A cDNA restores UV resistance to the mutant, indicating that rhp23 is a functional homolog of the human and S.cerevisiae genes. The rhp23::ura4 mutation also causes a delay in the G2 phase of the cell cycle which is corrected when rhp23, RAD23 or HHR23A cDNA is expressed. Rhp23 is present throughout the cell but is located predominantly in the nucleus, and the nuclear levels of Rhp23 decrease around the time of S phase in the cell cycle. Rhp23 is ubiquitinated at low levels, but overexpression of the rhp23 cDNA induces a large increase in ubiquitination of other proteins. Consistent with a role in protein ubiquitination, Rhp23 binds ubiquitin, as determined by two-hybrid analysis. Thus, the rhp23 gene plays a role not only in nucleotide excision repair but also in cell cycle regulation and the ubiquitination pathways.
Figures
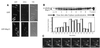


Similar articles
-
Identification and characterization of the rhp23(+) DNA repair gene in Schizosaccharomyces pombe.Biochem Biophys Res Commun. 2000 Feb 5;268(1):210-5. doi: 10.1006/bbrc.2000.2100. Biochem Biophys Res Commun. 2000. PMID: 10652237
-
Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance.Mol Cell Biol. 2003 Sep;23(18):6553-63. doi: 10.1128/MCB.23.18.6553-6563.2003. Mol Cell Biol. 2003. PMID: 12944481 Free PMC article.
-
Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23.Genomics. 1996 Jan 1;31(1):20-7. doi: 10.1006/geno.1996.0004. Genomics. 1996. PMID: 8808275
-
Repair of UV damage in the fission yeast Schizosaccharomyces pombe.Mutat Res. 2000 Jun 30;451(1-2):197-210. doi: 10.1016/s0027-5107(00)00050-6. Mutat Res. 2000. PMID: 10915873 Review.
-
The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation.DNA Repair (Amst). 2009 Apr 5;8(4):449-60. doi: 10.1016/j.dnarep.2009.01.005. Epub 2009 Feb 14. DNA Repair (Amst). 2009. PMID: 19223247 Review.
Cited by
-
APOBEC3G-UBA2 fusion as a potential strategy for stable expression of APOBEC3G and inhibition of HIV-1 replication.Retrovirology. 2008 Aug 4;5:72. doi: 10.1186/1742-4690-5-72. Retrovirology. 2008. PMID: 18680593 Free PMC article.
-
Structural determinants for the binding of ubiquitin-like domains to the proteasome.EMBO J. 2003 Sep 15;22(18):4634-45. doi: 10.1093/emboj/cdg467. EMBO J. 2003. PMID: 12970176 Free PMC article.
-
Photoprotective Role of Photolyase-Interacting RAD23 and Its Pleiotropic Effect on the Insect-Pathogenic Fungus Beauveria bassiana.Appl Environ Microbiol. 2020 May 19;86(11):e00287-20. doi: 10.1128/AEM.00287-20. Print 2020 May 19. Appl Environ Microbiol. 2020. PMID: 32245759 Free PMC article.
-
Vpr-host interactions during HIV-1 viral life cycle.J Neuroimmune Pharmacol. 2011 Jun;6(2):216-29. doi: 10.1007/s11481-011-9261-z. Epub 2011 Feb 12. J Neuroimmune Pharmacol. 2011. PMID: 21318276 Free PMC article. Review.
-
Lysine deserts prevent adventitious ubiquitylation of ubiquitin-proteasome components.Cell Mol Life Sci. 2023 May 9;80(6):143. doi: 10.1007/s00018-023-04782-z. Cell Mol Life Sci. 2023. PMID: 37160462 Free PMC article.
References
-
- Sturm A. and Lienhard,S. (1998) Two isoforms of plant RAD23 complement a UV-sensitive rad23 mutant in yeast. Plant J., 13, 815–821. - PubMed
-
- van der Spek P.J., Eker,A., Rademakers,S., Visser,C., Sugasawa,K., Masutani,C., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1996) XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res., 24, 2551–2559. - PMC - PubMed
-
- Tanaka K., Suzuki,T. and Chiba,T. (1998) The ligation systems for ubiquitin and ubiquitin-like proteins. Mol. Cell, 8, 503–512. - PubMed
-
- Hofmann K. and Bucher,P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

