The g5R (D250) gene of African swine fever virus encodes a Nudix hydrolase that preferentially degrades diphosphoinositol polyphosphates
- PMID: 11773415
- PMCID: PMC135849
- DOI: 10.1128/jvi.76.3.1415-1421.2002
The g5R (D250) gene of African swine fever virus encodes a Nudix hydrolase that preferentially degrades diphosphoinositol polyphosphates
Erratum in
- J Virol 2002 Jul;76(13):6864
Abstract
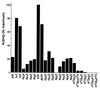
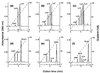
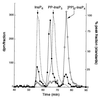
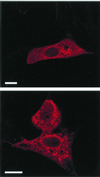
The African swine fever virus (ASFV) g5R gene encodes a protein containing a Nudix hydrolase motif which in terms of sequence appears most closely related to the mammalian diadenosine tetraphosphate (Ap4A) hydrolases. However, purified recombinant g5R protein (g5Rp) showed a much wider range of nucleotide substrate specificity compared to eukaryotic Ap4A hydrolases, having highest activity with GTP, followed by adenosine 5'-pentaphosphate (p5A) and dGTP. Diadenosine and diguanosine nucleotides were substrates, but the enzyme showed no activity with cap analogues such as 7mGp3A. In common with eukaryotic diadenosine hexaphosphate (Ap6A) hydrolases, which prefer higher-order polyphosphates as substrates, g5Rp also hydrolyzes the diphosphoinositol polyphosphates PP-InsP5 and [PP]2-InsP4. A comparison of the kinetics of substrate utilization showed that the k(cat)/K(m) ratio for PP-InsP5 is 60-fold higher than that for GTP, which allows classification of g5R as a novel diphosphoinositol polyphosphate phosphohydrolase (DIPP). Unlike mammalian DIPP, g5Rp appeared to preferentially remove the 5-beta-phosphate from both PP-InsP5 and [PP]2-InsP4. ASFV infection led to a reduction in the levels of PP-InsP5, ATP and GTP by ca. 50% at late times postinfection. The measured intracellular concentrations of these compounds were comparable to the respective K(m) values of g5Rp, suggesting that one or all of these may be substrates for g5Rp during ASFV infection. Transfection of ASFV-infected Vero cells with a plasmid encoding epitope-tagged g5Rp suggested localization of this protein in the rough endoplasmic reticulum. These results suggest a possible role for g5Rp in regulating a stage of viral morphogenesis involving diphosphoinositol polyphosphate-mediated membrane trafficking.
Figures


Similar articles
-
The African swine fever virus g5R protein possesses mRNA decapping activity.Virology. 2009 Oct 10;393(1):177-82. doi: 10.1016/j.virol.2009.07.026. Epub 2009 Aug 19. Virology. 2009. PMID: 19695654 Free PMC article.
-
Oxidation of the diphosphoinositol polyphosphate phosphohydrolase-like Nudix hydrolase Aps from Drosophila melanogaster induces thermolability--A possible regulatory switch?Int J Biochem Cell Biol. 2010 Jul;42(7):1174-81. doi: 10.1016/j.biocel.2010.04.003. Epub 2010 Apr 13. Int J Biochem Cell Biol. 2010. PMID: 20394834
-
Site-directed mutagenesis of diphosphoinositol polyphosphate phosphohydrolase, a dual specificity NUDT enzyme that attacks diadenosine polyphosphates and diphosphoinositol polyphosphates.J Biol Chem. 1999 Dec 10;274(50):35434-40. doi: 10.1074/jbc.274.50.35434. J Biol Chem. 1999. PMID: 10585413
-
The MutT motif family of nucleotide phosphohydrolases in man and human pathogens (review).Int J Mol Med. 1999 Jul;4(1):79-89. doi: 10.3892/ijmm.4.1.79. Int J Mol Med. 1999. PMID: 10373642 Review.
-
Structural insight into inositol pyrophosphate turnover.Adv Biol Regul. 2013 Jan;53(1):19-27. doi: 10.1016/j.jbior.2012.10.002. Epub 2012 Oct 11. Adv Biol Regul. 2013. PMID: 23107997 Free PMC article. Review.
Cited by
-
The African swine fever virus protein j4R binds to the alpha chain of nascent polypeptide-associated complex.J Virol. 2002 Oct;76(19):9991-9. doi: 10.1128/jvi.76.19.9991-9999.2002. J Virol. 2002. PMID: 12208975 Free PMC article.
-
Structure based prediction of functional sites with potential inhibitors to Nudix enzymes from disease causing microbes.Bioinformation. 2011 Jan 22;5(8):341-9. doi: 10.6026/97320630005341. Bioinformation. 2011. PMID: 21383922 Free PMC article.
-
African Swine Fever Virus Protein-Protein Interaction Prediction.Viruses. 2024 Jul 20;16(7):1170. doi: 10.3390/v16071170. Viruses. 2024. PMID: 39066332 Free PMC article.
-
Substrate specificity characterization for eight putative nudix hydrolases. Evaluation of criteria for substrate identification within the Nudix family.Proteins. 2016 Dec;84(12):1810-1822. doi: 10.1002/prot.25163. Epub 2016 Oct 1. Proteins. 2016. PMID: 27618147 Free PMC article.
-
Structure and function of African swine fever virus proteins: Current understanding.Front Microbiol. 2023 Feb 10;14:1043129. doi: 10.3389/fmicb.2023.1043129. eCollection 2023. Front Microbiol. 2023. PMID: 36846791 Free PMC article. Review.
References
-
- Bessman, M. J., D. N. Frick, and S. F. O’Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059–25062. - PubMed
-
- Caffrey, J. J., S. T. Safrany, X. N. Yang, and S. B. Shears. 2000. Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases: an expanding Nudt family. J. Biol. Chem. 275:12730–12736. - PubMed
-
- Cartwright, J. L., P. Britton, M. F. Minnick, and A. G. McLennan. 1999. The ialA invasion gene of Bartonella bacilliformis encodes a (di)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem. Biophys. Res. Commun. 256:474–479. - PubMed
-
- Cartwright, J. L., L. Gasmi, D. G. Spiller, and A. G. McLennan. 2000. The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal Nudix hydrolase active towards coenzyme A and its derivatives. J. Biol. Chem. 275:32925–32930. - PubMed
-
- Cartwright, J. L., and A. G. McLennan. 1999. The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5′,5′″-P1,P6-hexaphosphate hydrolase member of the MutT motif (Nudix hydrolase) family. J. Biol. Chem. 274:8604–8610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

