Retroviral vectors produced in the cytoplasmic vaccinia virus system transduce intron-containing genes
- PMID: 11773399
- PMCID: PMC135843
- DOI: 10.1128/jvi.76.3.1236-1243.2002
Retroviral vectors produced in the cytoplasmic vaccinia virus system transduce intron-containing genes
Abstract
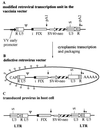
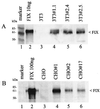
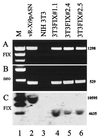
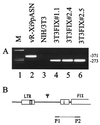
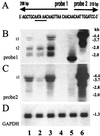
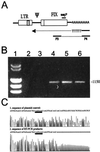
Introns and polyadenylation (pA) sites are known to improve transcript stability and nuclear-cytoplasmic transport and are normally present in efficient gene expression vectors. Standard retroviral vectors, however, do not allow the inclusion of such sequence elements, as mRNA processing at internal splice and pA sites interferes with the production of functional full-length vector genomes. In this report we examined the capability of hybrid vaccinia/retroviral vectors to transduce complex gene cassettes with nuclear RNA processing signals within the retroviral genome. A retroviral vector was constructed that contains a gene of interest (the human coagulation factor IX [FIX] cDNA), including an intron and an internal pA site. The modified proviral vector genome was cloned downstream of a vaccinia virus promoter and was inserted into the vaccinia virus genome. Infection of a packaging cell line with the recombinant vaccinia virus vector resulted in secretion of retroviral particles at average titers of 10(5) CFU per ml of cell culture supernatant. Due to the cytoplasmic transcription and the nonrecognition of nuclear transcription signals in the vaccinia virus system, full-length transcripts were obtained that still contained the intron. In the retrovirally transduced cell lines the FIX transcripts were terminated at the internal pA site. The transcripts were quantitatively spliced, and FIX was secreted. Recombinant cell lines with stable single-copy inserts containing sequence elements necessary for efficient gene function could be generated. Thus, a relatively simple cytoplasmic system for the generation of complex retroviral vectors is described. Retroviral vectors transducing intron-containing gene cassettes may play a further role in gene therapy applications.
Figures


Similar articles
-
Generation of transduction-competent retroviral vectors by infection with a single hybrid vaccinia virus.J Virol. 2003 Jun;77(12):7017-25. doi: 10.1128/jvi.77.12.7017-7025.2003. J Virol. 2003. PMID: 12768020 Free PMC article.
-
Vaccinia viral/retroviral chimeric vectors.Curr Gene Ther. 2004 Dec;4(4):417-26. doi: 10.2174/1566523043346101. Curr Gene Ther. 2004. PMID: 15578991 Review.
-
Poxviral/retroviral chimeric vectors allow cytoplasmic production of transducing defective retroviral particles.Virology. 1999 Jan 5;253(1):107-14. doi: 10.1006/viro.1998.9496. Virology. 1999. PMID: 9887323
-
Semliki Forest virus-mediated production of retroviral vector RNA in retroviral packaging cells.Hum Gene Ther. 1997 Nov 20;8(17):2031-41. doi: 10.1089/hum.1997.8.17-2031. Hum Gene Ther. 1997. PMID: 9414252
-
Retroviral Insertional Mutagenesis in Humans: Evidence for Four Genetic Mechanisms Promoting Expansion of Cell Clones.Mol Ther. 2020 Feb 5;28(2):352-356. doi: 10.1016/j.ymthe.2019.12.009. Epub 2020 Jan 7. Mol Ther. 2020. PMID: 31951833 Free PMC article. Review.
Cited by
-
Generation of transduction-competent retroviral vectors by infection with a single hybrid vaccinia virus.J Virol. 2003 Jun;77(12):7017-25. doi: 10.1128/jvi.77.12.7017-7025.2003. J Virol. 2003. PMID: 12768020 Free PMC article.
-
A vaccinia virus recombinant transcribing an alphavirus replicon and expressing alphavirus structural proteins leads to packaging of alphavirus infectious single cycle particles.PLoS One. 2013 Oct 9;8(10):e75574. doi: 10.1371/journal.pone.0075574. eCollection 2013. PLoS One. 2013. PMID: 24130722 Free PMC article.
-
Link between genome packaging and rate of budding for Rous sarcoma virus.J Virol. 2003 Sep;77(17):9388-98. doi: 10.1128/jvi.77.17.9388-9398.2003. J Virol. 2003. PMID: 12915554 Free PMC article.
-
Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome.J Virol. 2002 Nov;76(22):11518-29. doi: 10.1128/jvi.76.22.11518-11529.2002. J Virol. 2002. PMID: 12388713 Free PMC article.
References
-
- Abonour, R., D. A. Williams, L. Einhorn, K. M. Hall, J. Chen, J. Coffman, C. M. Traycoff, A. Bank, I. Kato, M. Ward, S. D. Williams, R. Hromas, M. J. Robertson, F. O. Smith, D. Woo, B. Mills, E. F. Srour, and K. Cornetta. 2000. Efficient retrovirus-mediated transfer of the multidrug resistance 1 gene into autologous human long-term repopulating hematopoietic stem cells. Nat. Med. 6:652–658. - PubMed
-
- Andreadis, S. T., C. M. Roth, J. M. Le Doux, J. R. Morgan, and M. L. Yarmush. 1999. Large-scale processing of recombinant retroviruses for gene therapy. Biotechnol. Prog. 15:1–11. - PubMed
-
- Barrett, P. N., A. Mitterer, W. Mundt, J. Eibl, M. Eibl, R. C. Gallo, B. Moss, and F. Dorner. 1989. Large-scale production and purification of a vaccinia recombinant-derived HIV-1 gp160 and analysis of its immunogenicity. AIDS Res. Hum. Retrovir. 5:159–171. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

