The p23 protein of citrus tristeza virus controls asymmetrical RNA accumulation
- PMID: 11752137
- PMCID: PMC136848
- DOI: 10.1128/jvi.76.2.473-483.2002
The p23 protein of citrus tristeza virus controls asymmetrical RNA accumulation
Abstract
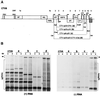
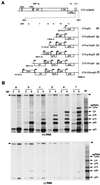
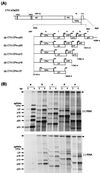
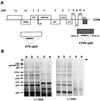
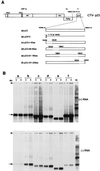
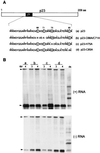
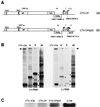
Citrus tristeza virus (CTV), a member of the Closteroviridae, has a 19.3-kb positive-stranded RNA genome that is organized into 12 open reading frames (ORFs) with the 10 3' genes expressed via a nested set of nine or ten 3'-coterminal subgenomic mRNAs (sgRNAs). Relatively large amounts of negative-stranded RNAs complementary to both genomic and sgRNAs accumulate in infected cells. As is characteristic of RNA viruses, wild-type CTV produced more positive than negative strands, with the plus-to-minus ratios of genomic and sgRNAs estimated at 10 to 20:1 and 40 to 50:1, respectively. However, a mutant with all of the 3' genes deleted replicated efficiently, but produced plus to minus strands at a markedly decreased ratio of 1 to 2:1. Deletion analysis of 3'-end genes revealed that the p23 ORF was involved in asymmetric RNA accumulation. A mutation which caused a frameshift after the fifth codon resulted in nearly symmetrical RNA accumulation, suggesting that the p23 protein, not a cis-acting element within the p23 ORF, controls asymmetric accumulation of CTV RNAs. Further in-frame deletion mutations in the p23 ORF suggested that amino acid residues 46 to 180, which contained RNA-binding and zinc finger domains, were indispensable for asymmetrical RNA accumulation, while the N-terminal 5 to 45 and C-terminal 181 to 209 amino acid residues were not absolutely required. Mutation of conserved cysteine residues to alanines in the zinc finger domain resulted in loss of activity of the p23 protein, suggesting involvement of the zinc finger in asymmetric RNA accumulation. The absence of p23 gene function was manifested by substantial increases in accumulation of negative-stranded RNAs and only modest decreases in positive-stranded RNAs. Moreover, the substantial decrease in the accumulation of negative-stranded coat protein (CP) sgRNA in the presence of the functional p23 gene resulted in a 12- to 15-fold increase in the expression of the CP gene. Apparently the excess negative-stranded sgRNA reduces the availability of the corresponding positive-stranded sgRNA as a messenger. Thus, the p23 protein controls asymmetric accumulation of CTV RNAs by downregulating negative-stranded RNA accumulation and indirectly increases expression of 3' genes.
Figures

Similar articles
-
Transcription strategy in a Closterovirus: a novel 5'-proximal controller element of Citrus Tristeza Virus produces 5'- and 3'-terminal subgenomic RNAs and differs from 3' open reading frame controller elements.J Virol. 2003 Jan;77(1):340-52. doi: 10.1128/jvi.77.1.340-352.2003. J Virol. 2003. PMID: 12477839 Free PMC article.
-
Characterization of the cis-acting elements controlling subgenomic mRNAs of citrus tristeza virus: production of positive- and negative-stranded 3'-terminal and positive-stranded 5'-terminal RNAs.Virology. 2001 Jul 20;286(1):134-51. doi: 10.1006/viro.2001.0987. Virology. 2001. PMID: 11448167
-
5'-coterminal subgenomic RNAs in citrus tristeza virus-infected cells.Virology. 2001 May 10;283(2):374-81. doi: 10.1006/viro.2001.0880. Virology. 2001. PMID: 11336562
-
Citrus tristeza virus: a pathogen that changed the course of the citrus industry.Mol Plant Pathol. 2008 Mar;9(2):251-68. doi: 10.1111/j.1364-3703.2007.00455.x. Mol Plant Pathol. 2008. PMID: 18705856 Free PMC article. Review.
-
From the smallest to the largest subcellular plant pathogen: Citrus tristeza virus and its unique p23 protein.Virus Res. 2022 Jun;314:198755. doi: 10.1016/j.virusres.2022.198755. Epub 2022 Mar 24. Virus Res. 2022. PMID: 35341876 Review.
Cited by
-
Citrus tristeza virus p23: a unique protein mediating key virus-host interactions.Front Microbiol. 2013 May 3;4:98. doi: 10.3389/fmicb.2013.00098. eCollection 2013. Front Microbiol. 2013. PMID: 23653624 Free PMC article.
-
Effects of modification of the transcription initiation site context on citrus tristeza virus subgenomic RNA synthesis.J Virol. 2003 Sep;77(17):9232-43. doi: 10.1128/jvi.77.17.9232-9243.2003. J Virol. 2003. PMID: 12915539 Free PMC article.
-
Protein-protein interactions between proteins of Citrus tristeza virus isolates.Virus Genes. 2014 Dec;49(3):456-65. doi: 10.1007/s11262-014-1100-x. Epub 2014 Jul 27. Virus Genes. 2014. PMID: 25064367
-
The p22 RNA silencing suppressor of the crinivirus Tomato chlorosis virus preferentially binds long dsRNAs preventing them from cleavage.Virology. 2016 Jan 15;488:129-36. doi: 10.1016/j.virol.2015.11.008. Epub 2015 Nov 27. Virology. 2016. PMID: 26629953 Free PMC article.
-
3'-coterminal subgenomic RNAs and putative cis-acting elements of Grapevine leafroll-associated virus 3 reveals 'unique' features of gene expression strategy in the genus Ampelovirus.Virol J. 2010 Aug 3;7:180. doi: 10.1186/1743-422X-7-180. Virol J. 2010. PMID: 20682046 Free PMC article.
References
-
- Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649–688. - PubMed
-
- Clarke, N. D., and J. M. Berg. 1998. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science 282:2018–2022. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

