A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69
- PMID: 11743003
- PMCID: PMC125785
- DOI: 10.1093/emboj/20.24.7271
A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69
Abstract
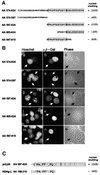
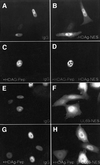
The best studied nuclear export processes are mediated by classical leucine-rich nuclear export signals that specify recognition by the CRM1 export receptor. However, details concerning alternative nuclear export signals and pathways are beginning to emerge. Within the family of Herpesviridae, a set of homologous regulatory proteins that are exemplified by the ICP27 of herpes simplex virus were described recently as nucleocytoplasmic shuttling proteins. Here we report that pUL69 of the beta-herpesvirus human cytomegalovirus is a nuclear protein that is able to shuttle between the nucleus and the cytoplasm independently of virus-encoded cofactors. In contrast to proteins containing a leucine-rich export signal, the shuttling activity of pUL69 was not affected by leptomycin B, indicating that pUL69 trafficking is not mediated by the export receptor CRM1. Importantly, we identified and characterized a novel type of transferable, leptomycin B-insensitive export signal that is distinct from other export signals described previously and is required for pUL69-mediated activation of gene expression. These data suggest that pUL69 is exported via a novel nuclear export pathway, based on a so far unique nuclear export signal of 28 amino acids.
Figures
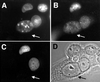
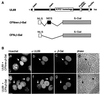

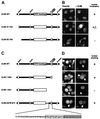


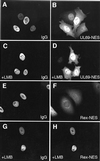


Similar articles
-
Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals.J Virol. 2006 Feb;80(3):1393-404. doi: 10.1128/JVI.80.3.1393-1404.2006. J Virol. 2006. PMID: 16415017 Free PMC article.
-
RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA.Nucleic Acids Res. 2006 Feb 25;34(4):1237-49. doi: 10.1093/nar/gkl007. Print 2006. Nucleic Acids Res. 2006. PMID: 16500893 Free PMC article.
-
The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA.Mol Cell Biol. 2006 Mar;26(5):1631-43. doi: 10.1128/MCB.26.5.1631-1643.2006. Mol Cell Biol. 2006. PMID: 16478985 Free PMC article.
-
Leucine-rich nuclear-export signals: born to be weak.Trends Cell Biol. 2005 Mar;15(3):121-4. doi: 10.1016/j.tcb.2005.01.005. Trends Cell Biol. 2005. PMID: 15752974 Review.
-
Interactions of human cytomegalovirus proteins with the nuclear transport machinery.Curr Top Microbiol Immunol. 2008;325:167-85. doi: 10.1007/978-3-540-77349-8_10. Curr Top Microbiol Immunol. 2008. PMID: 18637506 Review.
Cited by
-
Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals.J Virol. 2006 Feb;80(3):1393-404. doi: 10.1128/JVI.80.3.1393-1404.2006. J Virol. 2006. PMID: 16415017 Free PMC article.
-
Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation.Mol Cell Biol. 2002 Aug;22(16):5826-34. doi: 10.1128/MCB.22.16.5826-5834.2002. Mol Cell Biol. 2002. PMID: 12138193 Free PMC article.
-
Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses.Plant Physiol. 2009 Feb;149(2):760-74. doi: 10.1104/pp.108.132217. Epub 2008 Dec 3. Plant Physiol. 2009. PMID: 19052154 Free PMC article.
-
Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm.J Virol. 2006 Oct;80(20):10274-80. doi: 10.1128/JVI.00995-06. J Virol. 2006. PMID: 17005707 Free PMC article.
-
Snail nuclear transport: the gateways regulating epithelial-to-mesenchymal transition?Semin Cancer Biol. 2014 Aug;27:39-45. doi: 10.1016/j.semcancer.2014.06.003. Epub 2014 Jun 17. Semin Cancer Biol. 2014. PMID: 24954011 Free PMC article. Review.
References
-
- Andreoni M., Faircloth,M., Vugler,L. and Britt,W.J. (1989) A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods, 23, 157–167. - PubMed
-
- Bello L.J., Davison,A.J., Glenn,M.A., Whitehouse,A., Rethmeier,N., Schulz,T.F. and Barklie,C.J. (1999) The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol., 80, 3207–3215. - PubMed
-
- Brewer C.B. (1994) Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol., 43, 233–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

