Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication
- PMID: 11739696
- PMCID: PMC135711
- DOI: 10.1128/jvi.76.1.313-326.2002
Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication
Abstract
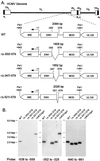
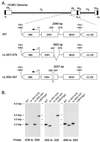
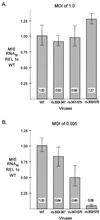
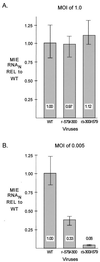
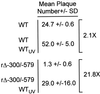
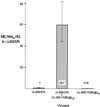
We have shown previously that the human cytomegalovirus (HCMV) major immediate-early (MIE) distal enhancer is needed for MIE promoter-dependent transcription and viral replication at low multiplicities of infection (MOI). To understand how this region works, we constructed and analyzed a series of HCMVs with various distal enhancer mutations. We show that the distal enhancer is composed of at least two parts that function independently to coordinately activate MIE promoter-dependent transcription and viral replication. One such part is contained in a 47-bp segment that has consensus binding sites for CREB/ATF, SP1, and YY1. At low MOI, these working parts likely function in cis to directly activate MIE gene expression, thus allowing viral replication to ensue. Three findings support the view that these working parts are likely cis-acting elements. (i) Deletion of either part of a bisegmented distal enhancer only slightly alters MIE gene transcription and viral replication. (ii) Reversing the distal enhancer's orientation largely preserves MIE gene transcription and viral replication. (iii) Placement of stop codons at -300 or -345 in all reading frames does not impair MIE gene transcription and viral replication. Lastly, we show that these working parts are dispensable at high MOI, partly because of compensatory stimulation of MIE promoter activity and viral replication that is induced by a virion-associated component(s) present at a high viral particle/cell ratio. We conclude that the distal enhancer is a complex multicomponent cis-acting region that is required to augment both MIE promoter-dependent transcription and HCMV replication.
Figures




Similar articles
-
The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection.J Virol. 2003 Mar;77(6):3602-14. doi: 10.1128/jvi.77.6.3602-3614.2003. J Virol. 2003. PMID: 12610136 Free PMC article.
-
The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression.J Virol. 2000 Feb;74(4):1602-13. doi: 10.1128/jvi.74.4.1602-1613.2000. J Virol. 2000. PMID: 10644329 Free PMC article.
-
A strong negative transcriptional regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer.J Virol. 1999 Nov;73(11):9039-52. doi: 10.1128/JVI.73.11.9039-9052.1999. J Virol. 1999. PMID: 10516010 Free PMC article.
-
Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency.Med Microbiol Immunol. 2008 Jun;197(2):223-31. doi: 10.1007/s00430-007-0069-7. Epub 2007 Dec 19. Med Microbiol Immunol. 2008. PMID: 18097687 Review.
-
Regulation of human cytomegalovirus immediate-early gene expression.Intervirology. 1996;39(5-6):331-42. doi: 10.1159/000150504. Intervirology. 1996. PMID: 9130043 Review.
Cited by
-
Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication.J Virol. 2005 Aug;79(15):9597-607. doi: 10.1128/JVI.79.15.9597-9607.2005. J Virol. 2005. PMID: 16014922 Free PMC article.
-
A BMPR2/YY1 Signaling Axis Is Required for Human Cytomegalovirus Latency in Undifferentiated Myeloid Cells.mBio. 2021 Jun 29;12(3):e0022721. doi: 10.1128/mBio.00227-21. Epub 2021 Jun 1. mBio. 2021. PMID: 34061599 Free PMC article.
-
The human cytomegalovirus UL133-138 gene locus attenuates the lytic viral cycle in fibroblasts.PLoS One. 2015 Mar 23;10(3):e0120946. doi: 10.1371/journal.pone.0120946. eCollection 2015. PLoS One. 2015. PMID: 25799165 Free PMC article.
-
Noncanonical TATA sequence in the UL44 late promoter of human cytomegalovirus is required for the accumulation of late viral transcripts.J Virol. 2008 Feb;82(4):1638-46. doi: 10.1128/JVI.01917-07. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057245 Free PMC article.
-
The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection.J Virol. 2003 Mar;77(6):3602-14. doi: 10.1128/jvi.77.6.3602-3614.2003. J Virol. 2003. PMID: 12610136 Free PMC article.
References
-
- Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within the human cytomegalovirus particles. Science 288:2373–2376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

