The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH
- PMID: 11713318
- PMCID: PMC92515
- DOI: 10.1093/nar/29.22.4684
The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH
Abstract
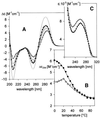
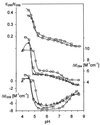
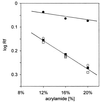
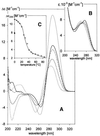
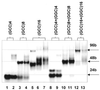
UV absorption and CD spectroscopy, along with polyacrylamide gel electrophoresis, were used to study conformational properties of DNA fragments containing the trinucleotide repeat (GCC)(n) (n = 4, 8 or 16), whose expansion is correlated with the fragile X chromosome syndrome. We have found that the conformational spectrum of the (GCC)(n) strand is wider than has been shown so far. (GCC)(n) strands adopt the hairpin described in the literature under a wide range of salt concentrations, but only at alkaline (>7.5) pH values. However, at neutral and slightly acid pH (GCC)(4) and (GCC)(8) strands homodimerize. Our data suggest that the homodimer is a bimolecular tetraplex formed by two parallel-oriented hairpins held together by hemi-protonated intermolecular C.C(+) pairs. The (GCC)(16) strand forms the same tetraplex intramolecularly. We further show that below pH 5 (GCC)(n) strands generate intercalated cytosine tetraplexes, whose molecularity depends on DNA strand length. They are tetramolecular with (GCC)(4), bimolecular with (GCC)(8) and monomolecular with (GCC)(16). i-Tetraplex formation is a complex and slow process. The neutral tetraplex, on the other hand, arises with fast kinetics under physiological conditions. Thus it is a conformational alternative of the (GCC)(n) strand duplex with a complementary (GGC)(n) strand.
Figures
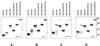

Similar articles
-
Conformational properties of DNA containing (CCA)n and (TGG)n trinucleotide repeats.Int J Biol Macromol. 2005 Jul;36(1-2):23-32. doi: 10.1016/j.ijbiomac.2005.03.005. Int J Biol Macromol. 2005. PMID: 15896838
-
The guanine-rich fragile X chromosome repeats are reluctant to form tetraplexes.Nucleic Acids Res. 2004 Jan 12;32(1):298-306. doi: 10.1093/nar/gkh179. Print 2004. Nucleic Acids Res. 2004. PMID: 14718550 Free PMC article.
-
Hairpin induced slippage and hyper-methylation of the fragile X DNA triplets.J Biomol Struct Dyn. 1998 Feb;15(4):745-56. doi: 10.1080/07391102.1998.10508989. J Biomol Struct Dyn. 1998. PMID: 9514250
-
Secondary structures in d(CGG) and d(CCG) repeat tracts.J Mol Biol. 1998 Jan 9;275(1):3-16. doi: 10.1006/jmbi.1997.1453. J Mol Biol. 1998. PMID: 9451434 Review.
-
Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X.J Mol Biol. 1994 Oct 21;243(2):143-51. doi: 10.1006/jmbi.1994.1640. J Mol Biol. 1994. PMID: 7932745 Review.
Cited by
-
Heterozygosity for a hypomorphic Polβ mutation reduces the expansion frequency in a mouse model of the Fragile X-related disorders.PLoS Genet. 2015 Apr 17;11(4):e1005181. doi: 10.1371/journal.pgen.1005181. eCollection 2015 Apr. PLoS Genet. 2015. PMID: 25886163 Free PMC article.
-
Advances in mechanisms of genetic instability related to hereditary neurological diseases.Nucleic Acids Res. 2005 Jul 8;33(12):3785-98. doi: 10.1093/nar/gki697. Print 2005. Nucleic Acids Res. 2005. PMID: 16006624 Free PMC article. Review.
-
Repeat-mediated genetic and epigenetic changes at the FMR1 locus in the Fragile X-related disorders.Front Genet. 2014 Jul 17;5:226. doi: 10.3389/fgene.2014.00226. eCollection 2014. Front Genet. 2014. PMID: 25101111 Free PMC article. Review.
-
Secondary structural choice of DNA and RNA associated with CGG/CCG trinucleotide repeat expansion rationalizes the RNA misprocessing in FXTAS.Sci Rep. 2021 Apr 14;11(1):8163. doi: 10.1038/s41598-021-87097-y. Sci Rep. 2021. PMID: 33854084 Free PMC article.
-
i-Motif DNA: structural features and significance to cell biology.Nucleic Acids Res. 2018 Sep 19;46(16):8038-8056. doi: 10.1093/nar/gky735. Nucleic Acids Res. 2018. PMID: 30124962 Free PMC article.
References
-
- Ashley C.T.J. and Warren,S.T. (1995) Trinucleotide repeat expansion and human disease. Annu. Rev. Genet., 29, 703–728. - PubMed
-
- Oostra B.A. and Willems,P.J. (1995) A fragile gene. Bioessays, 17, 941–947. - PubMed
-
- Jin P. and Warren,S.T. (2000) Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet., 9, 901–908. - PubMed
-
- Darlow J.M. and Leach,D.R.F. (1998) Secondary structures in d(CGG) and d(CCG) repeat tracts. J. Mol. Biol., 275, 3–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

