Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1
- PMID: 11711612
- PMCID: PMC116118
- DOI: 10.1128/JVI.75.24.12209-12219.2001
Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1
Abstract
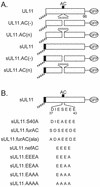
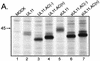
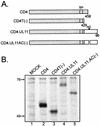
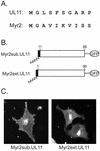
Growing evidence indicates that herpes simplex virus type 1 (HSV-1) acquires its final envelope in the trans-Golgi network (TGN). During the envelopment process, the viral nucleocapsid as well as the envelope and tegument proteins must arrive at this site in order to be incorporated into assembling virions. To gain a better understanding of how these proteins associate with cellular membranes and target to the correct compartment, we have been studying the intracellular trafficking properties of the small tegument protein encoded by the U(L)11 gene of HSV-1. This 96-amino-acid, myristylated protein accumulates on the cytoplasmic face of internal membranes, where it is thought to play a role in nucleocapsid envelopment and egress. When expressed in the absence of other HSV-1 proteins, the UL11 protein localizes to the Golgi apparatus, and previous deletion analyses have revealed that the membrane-trafficking information is contained within the first 49 amino acids. The goal of this study was to map the functional domains required for proper Golgi membrane localization. In addition to N-terminal myristylation, which allows for weak membrane binding, UL11 appears to be palmitylated on one or more of three consecutive N-terminal cysteines. Using membrane-pelleting experiments and confocal microscopy, we show that palmitylation of UL11 is required for both Golgi targeting specificity and strong membrane binding. Furthermore, we found that a conserved acidic cluster within the first half of UL11 is required for the recycling of this tegument protein from the plasma membrane to the Golgi apparatus. Taken together, our results demonstrate that UL11 has highly dynamic membrane-trafficking properties, which suggests that it may play multiple roles on the plasma membrane as well as on the nuclear and TGN membranes.
Figures









Similar articles
-
Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1.J Virol. 2006 Nov;80(21):10534-41. doi: 10.1128/JVI.01172-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928743 Free PMC article.
-
Binding partners for the UL11 tegument protein of herpes simplex virus type 1.J Virol. 2003 Nov;77(21):11417-24. doi: 10.1128/jvi.77.21.11417-11424.2003. J Virol. 2003. PMID: 14557627 Free PMC article.
-
UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment.J Virol. 2007 Apr;81(7):3097-108. doi: 10.1128/JVI.02201-06. Epub 2007 Jan 10. J Virol. 2007. PMID: 17215291 Free PMC article.
-
Virus Assembly and Egress of HSV.Adv Exp Med Biol. 2018;1045:23-44. doi: 10.1007/978-981-10-7230-7_2. Adv Exp Med Biol. 2018. PMID: 29896661 Review.
-
Functional roles of the tegument proteins of herpes simplex virus type 1.Virus Res. 2009 Nov;145(2):173-86. doi: 10.1016/j.virusres.2009.07.007. Epub 2009 Jul 15. Virus Res. 2009. PMID: 19615419 Review.
Cited by
-
HSV-1 Cytoplasmic Envelopment and Egress.Int J Mol Sci. 2020 Aug 19;21(17):5969. doi: 10.3390/ijms21175969. Int J Mol Sci. 2020. PMID: 32825127 Free PMC article. Review.
-
The amino terminus of the herpes simplex virus 1 protein Vhs mediates membrane association and tegument incorporation.J Virol. 2006 Oct;80(20):10117-27. doi: 10.1128/JVI.00744-06. J Virol. 2006. PMID: 17005689 Free PMC article.
-
Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus.J Virol. 2004 Jan;78(1):539-43. doi: 10.1128/jvi.78.1.539-543.2004. J Virol. 2004. PMID: 14671136 Free PMC article.
-
The C-Terminus of Epstein-Barr Virus BRRF2 Is Required for its Proper Localization and Efficient Virus Production.Front Microbiol. 2017 Jan 31;8:125. doi: 10.3389/fmicb.2017.00125. eCollection 2017. Front Microbiol. 2017. PMID: 28197146 Free PMC article.
-
Directional spread of alphaherpesviruses in the nervous system.Viruses. 2013 Feb 11;5(2):678-707. doi: 10.3390/v5020678. Viruses. 2013. PMID: 23435239 Free PMC article. Review.
References
-
- Berthiaume L, Resh M D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J Biol Chem. 1995;270:22399–22405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

