Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF
- PMID: 11701886
- PMCID: PMC139469
- DOI: 10.1105/tpc.010231
Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF
Abstract
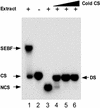
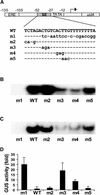
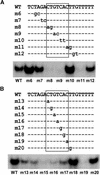
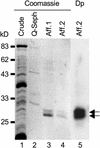
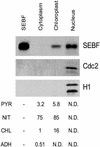
The potato pathogenesis-related gene PR-10a is transcriptionally activated in response to pathogen infection or elicitor treatment. Characterization of the cis-acting elements of the PR-10a promoter revealed the presence of a silencing element between residues -52 and -27 that contributes to transcriptional regulation. In this study, we have isolated a silencing element binding factor (SEBF) from potato tuber nuclei that binds to the coding strand of the silencing element in a sequence-specific manner. The consensus binding site of SEBF, PyTGTCNC, is present in a number of PR genes and shows striking similarity to the auxin response element. Mutational analysis of the PR-10a promoter revealed an inverse correlation between the in vitro binding of SEBF and the expression of PR-10a. SEBF was purified to homogeneity from potato tubers, and sequencing of the N terminus of the protein led to the isolation of a cDNA clone. Sequence analysis revealed that SEBF is homologous with chloroplast RNA binding proteins that possess consensus sequence-type RNA binding domains characteristic of heterogeneous nuclear ribonucleoproteins (hnRNPs). Overexpression of SEBF in protoplasts repressed the activity of a PR-10a reporter construct in a silencing element-dependent manner, confirming the role of SEBF as a transcriptional repressor.
Figures



Similar articles
-
The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene.Plant Cell. 2008 Nov;20(11):3136-47. doi: 10.1105/tpc.108.061721. Epub 2008 Nov 21. Plant Cell. 2008. PMID: 19028963 Free PMC article.
-
PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato.Plant Cell. 2000 Aug;12(8):1477-89. doi: 10.1105/tpc.12.8.1477. Plant Cell. 2000. PMID: 10948264 Free PMC article.
-
Over-expression of PR-10a leads to increased salt and osmotic tolerance in potato cell cultures.J Biotechnol. 2010 Nov;150(3):277-87. doi: 10.1016/j.jbiotec.2010.09.934. Epub 2010 Sep 17. J Biotechnol. 2010. PMID: 20854851
-
Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control.Plant Physiol. 2009 Aug;150(4):2030-44. doi: 10.1104/pp.109.140483. Epub 2009 Jun 3. Plant Physiol. 2009. PMID: 19493969 Free PMC article.
-
Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling.Mol Biol Rep. 2014 Feb;41(2):599-611. doi: 10.1007/s11033-013-2897-4. Epub 2013 Dec 17. Mol Biol Rep. 2014. PMID: 24343423 Review.
Cited by
-
Genome-Wide Analysis of LysM-Containing Gene Family in Wheat: Structural and Phylogenetic Analysis during Development and Defense.Genes (Basel). 2020 Dec 29;12(1):31. doi: 10.3390/genes12010031. Genes (Basel). 2020. PMID: 33383636 Free PMC article.
-
FaRE1: a transcriptionally active Ty1-copia retrotransposon in strawberry.J Plant Res. 2010 Sep;123(5):707-14. doi: 10.1007/s10265-009-0290-0. Epub 2009 Dec 18. J Plant Res. 2010. PMID: 20020171
-
Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis.Plant Physiol. 2009 Oct;151(2):843-56. doi: 10.1104/pp.109.140228. Epub 2009 Aug 5. Plant Physiol. 2009. PMID: 19656906 Free PMC article.
-
Three Pectin Methylesterase Inhibitors Protect Cell Wall Integrity for Arabidopsis Immunity to Botrytis.Plant Physiol. 2017 Mar;173(3):1844-1863. doi: 10.1104/pp.16.01185. Epub 2017 Jan 12. Plant Physiol. 2017. PMID: 28082716 Free PMC article.
-
Transcriptional activation and localization of expression of Brassica juncea putative metal transport protein BjMTP1.BMC Plant Biol. 2007 Jun 18;7:32. doi: 10.1186/1471-2229-7-32. BMC Plant Biol. 2007. PMID: 17577406 Free PMC article.
References
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2001). Current Protocols in Molecular Biology. (New York: Wiley).
-
- Blumwald, E., Gilad, S.A., and Lam, B.C.H. (1998). Early signal transduction pathways in plant–pathogen interactions. Trends Plant Sci. 3 342–346.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

