TissueInfo: high-throughput identification of tissue expression profiles and specificity
- PMID: 11691939
- PMCID: PMC60201
- DOI: 10.1093/nar/29.21.e102
TissueInfo: high-throughput identification of tissue expression profiles and specificity
Abstract
We describe TissueInfo, a knowledge-based method for the high-throughput identification of tissue expression profiles and tissue specificity. TissueInfo defines a set of tissue information calculations that can be computed for large numbers of genes, expressed sequence tags (ESTs) or proteins. Tissue information records that result from the TissueInfo calculations are used to generate tables suitable for data mining and for the selection of genes according to a given expression profile or specificity. When benchmarked against a test set of 116 proteins and literature information, TissueInfo was found to be accurate for 69% of identified tissue specificities and for 80% of expression profiles. The accuracy of the identifications can be increased if query sequences for which little information is available from dbEST are ignored. Thus, with 80% coverage, TissueInfo achieves an accuracy of 76% for specificity and 89% for expression. For the same set of proteins, the curated tissue specificity offered in SWISS-PROT was accurate in 78% of cases. TissueInfo can be useful for the selection of clones for custom microarrays, selection of training sets for ab initio identification of tissue information, gene discovery and genome-wide predictions. Further information about the program can be found at http://icb.mssm.edu/tissueinfo.
Figures
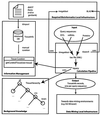
Similar articles
-
Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap and TissueInfo databases.BMC Genomics. 2003 Jul 29;4(1):31. doi: 10.1186/1471-2164-4-31. BMC Genomics. 2003. PMID: 12885301 Free PMC article.
-
Beyond tissueInfo: functional prediction using tissue expression profile similarity searches.Nucleic Acids Res. 2008 Jun;36(11):3728-37. doi: 10.1093/nar/gkn233. Epub 2008 May 15. Nucleic Acids Res. 2008. PMID: 18483083 Free PMC article.
-
HOMER: a human organ-specific molecular electronic repository.BMC Bioinformatics. 2011 Oct 18;12 Suppl 10(Suppl 10):S4. doi: 10.1186/1471-2105-12-S10-S4. BMC Bioinformatics. 2011. PMID: 22165817 Free PMC article.
-
Technology evaluation: SAGE, Genzyme molecular oncology.Curr Opin Mol Ther. 2001 Feb;3(1):85-96. Curr Opin Mol Ther. 2001. PMID: 11249736 Review.
-
Integration of bioinformatics resources for functional analysis of gene expression and proteomic data.Front Biosci. 2007 Sep 1;12:5071-88. doi: 10.2741/2449. Front Biosci. 2007. PMID: 17569631 Review.
Cited by
-
Whole-body distribution and metabolism of [N-methyl-11C](R)-1-(2-chlorophenyl)-N-(1-methylpropyl)-3-isoquinolinecarboxamide in humans; an imaging agent for in vivo assessment of peripheral benzodiazepine receptor activity with positron emission tomography.Eur J Nucl Med Mol Imaging. 2009 Apr;36(4):671-82. doi: 10.1007/s00259-008-1000-1. Epub 2008 Dec 3. Eur J Nucl Med Mol Imaging. 2009. PMID: 19050880
-
Quantitative gene expression profiles in real time from expressed sequence tag databases.Gene Expr. 2010;14(6):321-36. doi: 10.3727/105221610x12717040569820. Gene Expr. 2010. PMID: 20635574 Free PMC article.
-
CALHM1 deficiency impairs cerebral neuron activity and memory flexibility in mice.Sci Rep. 2016 Apr 12;6:24250. doi: 10.1038/srep24250. Sci Rep. 2016. PMID: 27066908 Free PMC article.
-
Effect of the CALHM1 G330D and R154H human variants on the control of cytosolic Ca2+ and Aβ levels.PLoS One. 2014 Nov 11;9(11):e112484. doi: 10.1371/journal.pone.0112484. eCollection 2014. PLoS One. 2014. PMID: 25386646 Free PMC article.
-
Systematic screening and identification of novel psoriasis‑specific genes from the transcriptome of psoriasis‑like keratinocytes.Mol Med Rep. 2019 Mar;19(3):1529-1542. doi: 10.3892/mmr.2018.9782. Epub 2018 Dec 20. Mol Med Rep. 2019. PMID: 30592269 Free PMC article.
References
-
- Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. - PubMed
-
- Marra M.A., Hillier,L. and Waterston,R.H. (1998) Expressed sequence tags—ESTablishing bridges between genomes. Trends Genet., 14, 4–7. - PubMed
-
- Putney S.D., Herlihy,W.C. and Schimmel,P. (1983) A new troponin T and cDNA clones for 13 different muscle proteins, found by shotgun sequencing. Nature, 302, 718–721. - PubMed
-
- Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

