Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta
- PMID: 11689430
- PMCID: PMC125708
- DOI: 10.1093/emboj/20.21.5898
Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta
Abstract
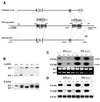
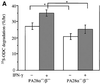
Two members of the proteasome activator, PA28alpha and PA28beta, form a heteropolymer that binds to both ends of the 20S proteasome. Evidence in vitro indicates that this interferon-gamma (IFN-gamma)-inducible heteropolymer is involved in the processing of intracellular antigens, but its functions in vivo remain elusive. To investigate the role of PA28alpha/beta in vivo, we generated mice deficient in both PA28alpha and PA28beta genes. The ATP-dependent proteolytic activities were decreased in PA28alpha(-/-)/beta(-/-) cells, suggesting that 'hybrid proteasomes' are involved in protein degradation. Treatment of PA28alpha(-/-)/beta(-/-) cells with IFN-gamma resulted in sufficient induction of the 'immunoproteasome'. Moreover, splenocytes from PA28alpha(-/-)/beta(-/-) mice displayed no apparent defects in processing of ovalbumin. These results are in marked contrast to the previous finding that immunoproteasome assembly and immune responses were impaired in PA28beta(-/-) mice. PA28alpha(-/-)/beta(-/-) mice also showed apparently normal immune responses against infection with influenza A virus. However, they almost completely lost the ability to process a melanoma antigen TRP2-derived peptide. Hence, PA28alpha/beta is not a prerequisite for antigen presentation in general, but plays an essential role for the processing of certain antigens.
Figures










Similar articles
-
A role for the proteasome regulator PA28alpha in antigen presentation.Nature. 1996 May 9;381(6578):166-8. doi: 10.1038/381166a0. Nature. 1996. PMID: 8610016
-
The proteasome regulator PA28alpha/beta can enhance antigen presentation without affecting 20S proteasome subunit composition.Eur J Immunol. 2000 Dec;30(12):3672-9. doi: 10.1002/1521-4141(200012)30:12<3672::AID-IMMU3672>3.0.CO;2-B. Eur J Immunol. 2000. PMID: 11169410
-
Expression of the proteasome activator PA28 rescues the presentation of a cytotoxic T lymphocyte epitope on melanoma cells.Cancer Res. 2002 May 15;62(10):2875-82. Cancer Res. 2002. PMID: 12019167
-
Structural plasticity of the proteasome and its function in antigen processing.Crit Rev Immunol. 2001;21(4):339-58. Crit Rev Immunol. 2001. PMID: 11922078 Review.
-
The proteasome activator 11 S REG (PA28) and class I antigen presentation.Biochem J. 2000 Jan 1;345 Pt 1(Pt 1):1-15. Biochem J. 2000. PMID: 10600633 Free PMC article. Review.
Cited by
-
Assembly of the 20S proteasome.Biochim Biophys Acta. 2014 Jan;1843(1):2-12. doi: 10.1016/j.bbamcr.2013.03.008. Epub 2013 Mar 16. Biochim Biophys Acta. 2014. PMID: 23507199 Free PMC article. Review.
-
Potential roles for PA28beta in gastric adenocarcinoma development and diagnosis.J Cancer Res Clin Oncol. 2010 Aug;136(8):1275-82. doi: 10.1007/s00432-010-0778-y. Epub 2010 Feb 6. J Cancer Res Clin Oncol. 2010. PMID: 20140627
-
A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans.J Clin Invest. 2011 Oct;121(10):4150-60. doi: 10.1172/JCI58414. Epub 2011 Sep 1. J Clin Invest. 2011. PMID: 21881205 Free PMC article.
-
PA28 and the proteasome immunosubunits play a central and independent role in the production of MHC class I-binding peptides in vivo.Eur J Immunol. 2011 Apr;41(4):926-35. doi: 10.1002/eji.201041040. Epub 2011 Mar 1. Eur J Immunol. 2011. PMID: 21360704 Free PMC article.
-
In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals.Biomolecules. 2019 May 31;9(6):213. doi: 10.3390/biom9060213. Biomolecules. 2019. PMID: 31159305 Free PMC article.
References
-
- Ben-Shahar S., Komlosh,A., Nadav,E., Shaked,I., Ziv,T., Admon,A., DeMartino,G.N. and Reiss,Y. (1999) 26S proteasome-mediated production of an authentic major histocompatibility class I-restricted epitope from an intact protein substrate. J. Biol. Chem., 274, 21963–21972. - PubMed
-
- Bochtler M., Ditzel,L., Groll,M., Hartmann,C. and Huber,R. (1999) The proteasome. Annu. Rev. Biophys. Biomol. Struct., 28, 295–317. - PubMed
-
- Coux O., Tanaka,K. and Goldberg,A.L. (1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem., 65, 801–847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

