The notch intracellular domain can function as a coactivator for LEF-1
- PMID: 11604490
- PMCID: PMC99925
- DOI: 10.1128/MCB.21.22.7537-7544.2001
The notch intracellular domain can function as a coactivator for LEF-1
Abstract
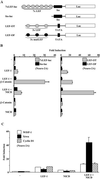
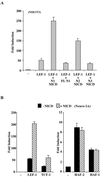
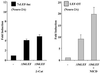
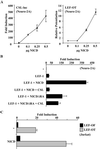
Notch signaling commences with two ligand-mediated proteolysis events that release the Notch intracellular domain, NICD, from the plasma membrane. NICD then translocates into the nucleus and interacts with the DNA binding protein CSL to activate transcription. We found that NICD expression also potentiates activity of the transcription factor LEF-1. NICD stimulation of LEF-1 activity was context dependent and occurred on a subset of promoters distinct from those activated by beta-catenin. Importantly, the effect of NICD does not appear to be mediated through canonical components of the Wnt signaling pathway or downstream components of the Notch pathway. In vitro assays show a weak association between the C-terminal transactivation domain of NICD and the high-mobility group domain of LEF-1, suggesting that the two proteins interact in vivo. Our data therefore describe a new nuclear target of Notch signaling and a new coactivator for LEF-1.
Figures



Similar articles
-
Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro.Genes Dev. 2001 Dec 15;15(24):3342-54. doi: 10.1101/gad.946501. Genes Dev. 2001. PMID: 11751639 Free PMC article.
-
Beta-catenin can act as a nuclear import receptor for its partner transcription factor, lymphocyte enhancer factor-1 (lef-1).Exp Cell Res. 2005 Aug 15;308(2):357-63. doi: 10.1016/j.yexcr.2005.05.011. Exp Cell Res. 2005. PMID: 15936755
-
Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1.EMBO J. 2001 Aug 15;20(16):4500-11. doi: 10.1093/emboj/20.16.4500. EMBO J. 2001. PMID: 11500377 Free PMC article.
-
Regulating the regulators: routing the Wnt-beta-catenin--Lef signals.J Invest Dermatol. 2004 Aug;123(2):VIII-X. doi: 10.1111/j.0022-202X.2004.23239.x. J Invest Dermatol. 2004. PMID: 15245452 Review. No abstract available.
-
CSL-independent Notch signalling: a checkpoint in cell fate decisions during development?Curr Opin Genet Dev. 2002 Oct;12(5):524-33. doi: 10.1016/s0959-437x(02)00336-2. Curr Opin Genet Dev. 2002. PMID: 12200157 Review.
Cited by
-
The Notch-1 intracellular domain is found in sub-nuclear bodies in SH-SY5Y neuroblastomas and in primary cortical neurons.Neurosci Lett. 2007 Mar 26;415(2):135-9. doi: 10.1016/j.neulet.2007.01.049. Epub 2007 Jan 27. Neurosci Lett. 2007. PMID: 17300869 Free PMC article.
-
Ligand-independent traffic of Notch buffers activated Armadillo in Drosophila.PLoS Biol. 2009 Aug;7(8):e1000169. doi: 10.1371/journal.pbio.1000169. Epub 2009 Aug 11. PLoS Biol. 2009. PMID: 19668359 Free PMC article.
-
Wnt-Notch signalling crosstalk in development and disease.Cell Mol Life Sci. 2014 Sep;71(18):3553-67. doi: 10.1007/s00018-014-1644-x. Epub 2014 Jun 19. Cell Mol Life Sci. 2014. PMID: 24942883 Free PMC article. Review.
-
Notch signalling pathway in tooth development and adult dental cells.Cell Prolif. 2011 Dec;44(6):495-507. doi: 10.1111/j.1365-2184.2011.00780.x. Epub 2011 Oct 4. Cell Prolif. 2011. PMID: 21973022 Free PMC article. Review.
-
Notch and Wnt signaling, physiological stimuli and postnatal myogenesis.Int J Biol Sci. 2010 May 15;6(3):268-81. doi: 10.7150/ijbs.6.268. Int J Biol Sci. 2010. PMID: 20567496 Free PMC article. Review.
References
-
- Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
-
- Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. - PubMed
-
- Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. - PubMed
-
- Beckman H, Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991;5:1057–1066. - PubMed
-
- Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
