Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression
- PMID: 11602710
- PMCID: PMC114650
- DOI: 10.1128/JVI.75.22.10683-10695.2001
Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression
Abstract
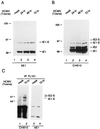
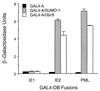
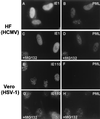
Human cytomegalovirus (HCMV) major immediate-early protein IE1 is an abundant 72-kDa nuclear phosphoprotein that is thought to play an important role in efficient triggering of the lytic cycle, especially at low multiplicity of infection. The best-known properties of IE1 at present are its transient targeting to punctate promyelocytic leukemia protein (PML)-associated nuclear bodies (PML oncogenic domains [PODs] or nuclear domain 10 [ND10]), with associated displacement of the cellular PML tumor suppressor protein into a diffuse nucleoplasmic form and its association with metaphase chromosomes. Recent studies have shown that the targeting of PML (and associated proteins such as hDaxx) to PODs is dependent on modification of PML by ubiquitin-like protein SUMO-1. In this study, we provide direct evidence that IE1 is also covalently modified by SUMO-1 in both infected and cotransfected cells, as well as in in vitro assays, with up to 30% of the protein representing the covalently conjugated 90-kDa form in stable U373/IE1 cell lines. Lysine 450 was mapped as the major SUMO-1 conjugation site, but a point mutation of this lysine residue in IE1 did not interfere with its targeting to and disruption of the PODs. Surprisingly, unlike PML or IE2, IE1 did not interact with either Ubc9 or SUMO-1 in yeast two-hybrid assays, suggesting that some additional unknown intranuclear cofactors must play a role in IE1 sumoylation. Interestingly, stable expression of either exogenous PML or exogenous Flag-SUMO-1 in U373 cell lines greatly enhanced both the levels and rate of in vivo IE1 sumoylation during HCMV infection. Unlike the disruption of PODs by the herpes simplex virus type 1 IE110(ICP0) protein, the disruption of PODs by HCMV IE1 proved not to involve proteasome-dependent degradation of PML. We also demonstrate here that the 560-amino-acid PML1 isoform functions as a transcriptional repressor when fused to the GAL4 DNA-binding domain and that wild-type IE1 inhibits the repressor function of PML1 in transient cotransfection assays. Furthermore, both IE1(1-346) and IE1(L174P) mutants, which are defective in displacing PML from PODs, failed to inhibit the repression activity of PML1, whereas the sumoylation-negative IE1(K450R) mutant derepressed as efficiently as wild-type IE1. Taken together, our results suggest that proteasome-independent disruption of PODs, but not IE1 sumoylation, is required for efficient IE1 inhibition of PML-mediated transcriptional repression.
Figures

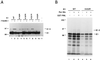


Similar articles
-
Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells.J Virol. 2004 Jun;78(12):6527-42. doi: 10.1128/JVI.78.12.6527-6542.2004. J Virol. 2004. PMID: 15163746 Free PMC article.
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells.J Virol. 1997 Jun;71(6):4599-613. doi: 10.1128/JVI.71.6.4599-4613.1997. J Virol. 1997. PMID: 9151854 Free PMC article.
-
PML and the oncogenic nuclear domains in regulating transcriptional repression.Curr Opin Cell Biol. 2000 Oct;12(5):641-4. doi: 10.1016/s0955-0674(00)00144-7. Curr Opin Cell Biol. 2000. PMID: 10978902 Review.
-
The Human CMV IE1 Protein: An Offender of PML Nuclear Bodies.Adv Anat Embryol Cell Biol. 2017;223:77-94. doi: 10.1007/978-3-319-53168-7_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28528440 Review.
Cited by
-
Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response.PLoS Pathog. 2016 Jul 7;12(7):e1005748. doi: 10.1371/journal.ppat.1005748. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387064 Free PMC article.
-
Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha.Proc Natl Acad Sci U S A. 2002 Aug 6;99(16):10683-8. doi: 10.1073/pnas.162352299. Epub 2002 Jul 26. Proc Natl Acad Sci U S A. 2002. PMID: 12145325 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification.J Virol. 2005 Aug;79(15):9912-25. doi: 10.1128/JVI.79.15.9912-9925.2005. J Virol. 2005. PMID: 16014952 Free PMC article.
-
The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling.Viruses. 2014 Apr 2;6(4):1502-24. doi: 10.3390/v6041502. Viruses. 2014. PMID: 24699362 Free PMC article.
-
Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO.J Virol. 2008 Nov;82(21):10444-54. doi: 10.1128/JVI.00833-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701593 Free PMC article.
References
-
- Ahn J H, Chiou C J, Hayward G S. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene. 1998;210:25–36. - PubMed
-
- Ahn J H, Hayward G S. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

