Protein-protein interaction panel using mouse full-length cDNAs
- PMID: 11591653
- PMCID: PMC311163
- DOI: 10.1101/gr.180101
Protein-protein interaction panel using mouse full-length cDNAs
Abstract
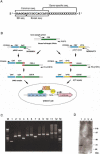
We have developed a novel assay system for systematic analysis of protein-protein interactions (PPIs) that is characteristic of a PCR-mediated rapid sample preparation and a high-throughput assay system based on the mammalian two-hybrid method. Using gene-specific primers, we successfully constructed the assay samples by two rounds of PCR with up to 3.6 kb from the first-round PCR fragments. In the assay system, we designed all the steps to be performed by adding only samples, reagents, and cells into 384-well assay plates using two types of semiautomatic multiple dispensers. The system enabled us examine more than 20,000 assay wells per day. We detected 145 interactions in our pilot study using 3500 samples derived from mouse full-length enriched cDNAs. Analysis of the interaction data showed both several significant interaction clusters and predicted functions of a few uncharacterized proteins. In combination with our comprehensive mouse full-length cDNA clone bank covering a large part of the whole genes, our high-throughput assay system will discover many interactions to facilitate understanding of the function of uncharacterized proteins and the molecular mechanism of crucial biological processes, and also enable completion of a rough draft of the entire PPI panel in certain cell types or tissues of mouse within a short time.
Figures

Similar articles
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D502-4. doi: 10.1093/nar/gkh035. Nucleic Acids Res. 2004. PMID: 14681467 Free PMC article.
-
Identification of promethin and PGLP as two novel up-regulated genes in PPARgamma1-induced adipogenic mouse liver.Biochimie. 2004 Nov;86(11):743-61. doi: 10.1016/j.biochi.2004.09.015. Biochimie. 2004. PMID: 15589683
-
High-throughput two-hybrid analysis. The promise and the peril.FEBS J. 2005 Nov;272(21):5391-9. doi: 10.1111/j.1742-4658.2005.04973.x. FEBS J. 2005. PMID: 16262681 Review.
-
[Problems and limitations of conventional and innovative methods for the diagnosis of Toxoplasmosis in humans and animals].Parassitologia. 2004 Jun;46(1-2):177-81. Parassitologia. 2004. PMID: 15305712 Review. Italian.
Cited by
-
Linking the Neuropsychiatric Disease Gene TCF4 to Neuronal Activity-Dependent Regulatory Networks.J Neurosci. 2018 Mar 14;38(11):2653-2655. doi: 10.1523/JNEUROSCI.3475-17.2018. J Neurosci. 2018. PMID: 29540544 Free PMC article. No abstract available.
-
Mis-targeting of the mitochondrial protein LIPT2 leads to apoptotic cell death.PLoS One. 2017 Jun 19;12(6):e0179591. doi: 10.1371/journal.pone.0179591. eCollection 2017. PLoS One. 2017. PMID: 28628643 Free PMC article.
-
The mammalian protein-protein interaction database and its viewing system that is linked to the main FANTOM2 viewer.Genome Res. 2003 Jun;13(6B):1534-41. doi: 10.1101/gr.956303. Genome Res. 2003. PMID: 12819152 Free PMC article.
-
Development of a high-throughput method for the systematic identification of human proteins nuclear translocation potential.BMC Cell Biol. 2009 Sep 22;10:69. doi: 10.1186/1471-2121-10-69. BMC Cell Biol. 2009. PMID: 19772597 Free PMC article.
-
A Novel Terminator Primer and Enhancer Reagents for Direct Expression of PCR-Amplified Genes in Mammalian Cells.Mol Biotechnol. 2015 Aug;57(8):767-80. doi: 10.1007/s12033-015-9870-5. Mol Biotechnol. 2015. PMID: 25997599
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Cochran AG. Antagonists of protein–protein interactions. Chem Biol. 2000;7:R85–R94. - PubMed
-
- Colas P, Brent R. The impact of two-hybrid and related methods on biotechnology. Trends Biotechnol. 1998;16:355–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
