Hydrogen-bonding classes in proteins and their contribution to the unfolding reaction
- PMID: 11567098
- PMCID: PMC2374212
- DOI: 10.1110/ps.09201
Hydrogen-bonding classes in proteins and their contribution to the unfolding reaction
Abstract
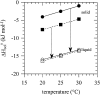
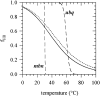
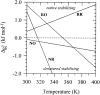
This paper proposes to assess hydrogen-bonding contributions to the protein stability, using a set of model proteins for which both X-ray structures and calorimetric unfolding data are known. Pertinent thermodynamic quantities are first estimated according to a recent model of protein energetics based on the dissolution of alkyl amides. Then it is shown that the overall free energy of hydrogen-bond formation accounts for a hydrogen-bonding propensity close to helix-forming tendencies previously found for individual amino acids. This allows us to simulate the melting curve of an alanine-rich helical 50-mer with good precision. Thereafter, hydrogen-bonding enthalpies and entropies are expressed as linear combinations of backbone-backbone, backbone-side-chain, side-chain-backbone, and side-chain-side-chain donor-acceptor contributions. On this basis, each of the four components shows a different free energy versus temperature trend. It appears that structural preference for side-chain-side-chain hydrogen bonding plays a major role in stabilizing proteins at elevated temperatures.
Figures
Similar articles
-
A calorimetric study of the folding-unfolding of an alpha-helix with covalently closed N and C-terminal loops.J Mol Biol. 1999 Aug 27;291(4):965-76. doi: 10.1006/jmbi.1999.3025. J Mol Biol. 1999. PMID: 10452900
-
Contribution of hydration to protein folding thermodynamics. I. The enthalpy of hydration.J Mol Biol. 1993 Jul 20;232(2):639-59. doi: 10.1006/jmbi.1993.1416. J Mol Biol. 1993. PMID: 8393940
-
Direct analysis of backbone-backbone hydrogen bond formation in protein folding transition states.J Mol Biol. 2006 Oct 20;363(2):506-19. doi: 10.1016/j.jmb.2006.07.058. Epub 2006 Jul 29. J Mol Biol. 2006. PMID: 16963082
-
Structure and energetics of the hydrogen-bonded backbone in protein folding.Annu Rev Biochem. 2008;77:339-62. doi: 10.1146/annurev.biochem.77.061306.131357. Annu Rev Biochem. 2008. PMID: 18518824 Review.
-
Backbone-Backbone H-Bonds Make Context-Dependent Contributions to Protein Folding Kinetics and Thermodynamics: Lessons from Amide-to-Ester Mutations.Adv Protein Chem. 2005;72:39-78. doi: 10.1016/S0065-3233(05)72002-7. Adv Protein Chem. 2005. PMID: 16581372 Review.
Cited by
-
Exploring the Structural Mechanism of Covalently Bound E3 Ubiquitin Ligase: Catalytic or Allosteric Inhibition?Protein J. 2018 Dec;37(6):500-509. doi: 10.1007/s10930-018-9795-5. Protein J. 2018. PMID: 30232697
-
Molecular Dynamic Simulation of Space and Earth-Grown Crystal Structures of Thermostable T1 Lipase Geobacillus zalihae Revealed a Better Structure.Molecules. 2017 Sep 25;22(10):1574. doi: 10.3390/molecules22101574. Molecules. 2017. PMID: 28946656 Free PMC article.
-
A multi-factors rational design strategy for enhancing the thermostability of Escherichia coli AppA phytase.J Ind Microbiol Biotechnol. 2013 May;40(5):457-64. doi: 10.1007/s10295-013-1260-z. Epub 2013 Mar 14. J Ind Microbiol Biotechnol. 2013. PMID: 23494709
-
Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion.Int J Mol Sci. 2019 May 24;20(10):2561. doi: 10.3390/ijms20102561. Int J Mol Sci. 2019. PMID: 31137725 Free PMC article.
-
Contribution of charged groups to the enthalpic stabilization of the folded states of globular proteins.J Phys Chem B. 2008 May 15;112(19):6159-67. doi: 10.1021/jp077024d. Epub 2008 Feb 28. J Phys Chem B. 2008. PMID: 18303881 Free PMC article.
References
-
- Becktel, W.J. and Schellman, J.A. 1987. Protein stability curves. Biopolymers 26 1859–1877. - PubMed
-
- Chen, C.-C., Zhu, Y., King, J.A., and Evans, L.B. 1992. A molecular thermodynamic approach to predict the secondary structure of homopolypeptides in aqueous systems. Biopolymers 32 1375–1392. - PubMed
-
- Costas, M., Kronberg, B., and Silveston, R. 1994. General thermodynamic analysis of the dissolution of non-polar molecules into water. J. Chem. Soc., Faraday Trans. 90 1513–1522.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

