Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells
- PMID: 11557811
- PMCID: PMC55920
- DOI: 10.1093/nar/29.18.3796
Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells
Abstract
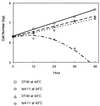
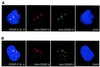
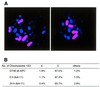
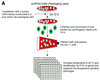
CENP-C is an evolutionarily conserved centromere protein that is thought to be an important component in kinetochore assembly in vertebrate cells. However, the functional role of CENP-C in cell cycle progression remains unclear. To further understand CENP-C function, we developed a method incorporating the hyper-recombinogenic chicken B lymphocyte cell line DT40 to create several temperature-sensitive CENP-C mutants in DT40 cells. We found that, under restrictive conditions, one temperature-sensitive mutant, ts4-11, displayed metaphase delay and chromosome missegregation but proceeded through the cell cycle until arrest at G(1) phase. Furthermore, ts4-11 cells were transfected with a human HeLa cell cDNA library maintained in a retroviral vector, and genes that suppressed the temperature-sensitive phenotype were identified. One of these suppressor genes encodes SUMO-1, which is a ubiquitin-like protein. This finding suggests that SUMO-1 may be involved in centromere function in vertebrate cells. The novel strategy reported here will be useful and applicable to a wide range of proteins that have general cell-autonomous function in vertebrate cells.
Figures





Similar articles
-
CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells.EMBO J. 2001 Aug 15;20(16):4603-17. doi: 10.1093/emboj/20.16.4603. EMBO J. 2001. PMID: 11500386 Free PMC article.
-
Efficient conditional mutation of the vertebrate CENP-C gene.Hum Mol Genet. 1997 Dec;6(13):2301-8. doi: 10.1093/hmg/6.13.2301. Hum Mol Genet. 1997. PMID: 9361037
-
The constitutive centromere component CENP-50 is required for recovery from spindle damage.Mol Cell Biol. 2005 Dec;25(23):10315-28. doi: 10.1128/MCB.25.23.10315-10328.2005. Mol Cell Biol. 2005. PMID: 16287847 Free PMC article.
-
The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres.Chromosome Res. 2004;12(6):543-56. doi: 10.1023/B:CHRO.0000036593.72788.99. Chromosome Res. 2004. PMID: 15289662 Review.
-
Let's think again about using mammalian temperature-sensitive mutants to investigate functional molecules-The perspectives from the studies on three mutants showing chromosome instability.J Cell Biochem. 2018 Sep;119(9):7143-7150. doi: 10.1002/jcb.27205. Epub 2018 Jun 26. J Cell Biochem. 2018. PMID: 29943840 Review.
Cited by
-
Characterization of a temperature-sensitive vertebrate clathrin heavy chain mutant as a tool to study clathrin-dependent events in vivo.PLoS One. 2010 Aug 6;5(8):e12017. doi: 10.1371/journal.pone.0012017. PLoS One. 2010. PMID: 20700507 Free PMC article.
-
Recruitment of the Ulp2 protease to the inner kinetochore prevents its hyper-sumoylation to ensure accurate chromosome segregation.PLoS Genet. 2019 Nov 20;15(11):e1008477. doi: 10.1371/journal.pgen.1008477. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31747400 Free PMC article.
-
Centromeres: unique chromatin structures that drive chromosome segregation.Nat Rev Mol Cell Biol. 2011 May;12(5):320-32. doi: 10.1038/nrm3107. Nat Rev Mol Cell Biol. 2011. PMID: 21508988 Free PMC article. Review.
-
Transcription in the maintenance of centromere chromatin identity.Nucleic Acids Res. 2012 Dec;40(22):11178-88. doi: 10.1093/nar/gks921. Epub 2012 Oct 11. Nucleic Acids Res. 2012. PMID: 23066104 Free PMC article. Review.
-
The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase.Cell Cycle. 2010 Aug 15;9(16):3194-201. doi: 10.4161/cc.9.16.12619. Epub 2010 Aug 9. Cell Cycle. 2010. PMID: 20724819 Free PMC article. Review.
References
-
- Choo K.H.A. (1997) The Centromere. Oxford University Press, Oxford, New York, Tokyo.
-
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. - PubMed
-
- Pidoux A.L. and Allshire,R.C. (2000) Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol., 12, 308–319. - PubMed
-
- Stoler S., Keith,K.C., Curnick,K.E. and Fitzgerald-Hayes,M. (1995) A mutation in CSE4, an essential gene encoding a novel chromatin associated protein in yeast, cause chromosomes nondisjunction and cell cycle arrest at mitosis. Genes Dev., 9, 573–586. - PubMed

