Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design
- PMID: 11533184
- PMCID: PMC114489
- DOI: 10.1128/JVI.75.19.9210-9228.2001
Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design
Abstract
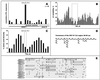
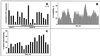
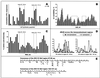
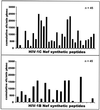
The most severe human immunodeficiency virus type 1 (HIV-1) epidemic is occurring in southern Africa. It is caused by HIV-1 subtype C (HIV-1C). In this study we present the identification and analysis of cumulative cytotoxic T-lymphocyte (CTL) responses in the southern African country of Botswana. CTLs were shown to be an important component of the immune response to control HIV-1 infection. The definition of optimal and dominant epitopes across the HIV-1C genome that are targeted by CTL is critical for vaccine design. The characteristics of the predominant virus that causes the HIV-1 epidemic in a certain geographic area and also the genetic background of the population, through the distribution of common HLA class I alleles, might impact dominant CTL responses in the vaccinee and in the general population. The enzyme-linked immunospot (Elispot) gamma interferon assay has recently been shown to be a reliable tool to map optimal CTL epitopes, correlating well with other methods, such as intracellular staining, tetramer staining, and the classical chromium release assay. Using Elispot with overlapping synthetic peptides across Gag, Tat, Rev, and Nef, we analyzed HIV-1C-specific CTL responses of HIV-1-infected blood donors. Profiles of cumulative Elispot-based CTL responses combined with diversity and sequence consensus data provide an additional characterization of immunodominant regions across the HIV-1C genome. Results of the study suggest that the construction of a poly-epitope subtype-specific HIV-1 vaccine that includes multiple copies of immunodominant CTL epitopes across the viral genome, derived from predominant HIV-1 viruses, might be a logical approach to the design of a vaccine against AIDS.
Figures






Similar articles
-
Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef.J Infect Dis. 2005 Sep 1;192(5):749-59. doi: 10.1086/432547. Epub 2005 Jul 27. J Infect Dis. 2005. PMID: 16088824
-
[Specific immune responses to human immunodeficiency virus type 1 Gag, Tat, Rev and Nef proteins].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2005 Mar;21(2):175-9. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2005. PMID: 15766402 Chinese.
-
Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C.J Virol. 2002 Oct;76(20):10155-68. doi: 10.1128/jvi.76.20.10155-10168.2002. J Virol. 2002. PMID: 12239290 Free PMC article.
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
-
Regulatory and accessory HIV-1 proteins: potential targets for HIV-1 vaccines?Curr Med Chem. 2005;12(6):741-7. doi: 10.2174/0929867053202205. Curr Med Chem. 2005. PMID: 15790309 Review.
Cited by
-
Human immunodeficiency virus type 1 gag-specific mucosal immunity after oral immunization with papillomavirus pseudoviruses encoding gag.J Virol. 2004 Oct;78(19):10249-57. doi: 10.1128/JVI.78.19.10249-10257.2004. J Virol. 2004. PMID: 15367590 Free PMC article.
-
An integrative bioinformatic approach for studying escape mutations in human immunodeficiency virus type 1 gag in the Pumwani Sex Worker Cohort.J Virol. 2008 Feb;82(4):1980-92. doi: 10.1128/JVI.02742-06. Epub 2007 Dec 5. J Virol. 2008. PMID: 18057233 Free PMC article.
-
Recombinant Salmonella enterica serovar Typhimurium as a vaccine vector for HIV-1 Gag.Viruses. 2013 Aug 28;5(9):2062-78. doi: 10.3390/v5092062. Viruses. 2013. PMID: 23989890 Free PMC article. Review.
-
Broad and persistent Gag-specific CD8+ T-cell responses are associated with viral control but rarely drive viral escape during primary HIV-1 infection.AIDS. 2015 Jan 2;29(1):23-33. doi: 10.1097/QAD.0000000000000508. AIDS. 2015. PMID: 25387316 Free PMC article.
-
Major histocompatibility complex class II (HLA-DRB and -DQB) allele frequencies in Botswana: association with human immunodeficiency virus type 1 infection.Clin Diagn Lab Immunol. 2005 Sep;12(9):1020-8. doi: 10.1128/CDLI.12.9.1020-1028.2005. Clin Diagn Lab Immunol. 2005. PMID: 16148166 Free PMC article.
References
-
- Abebe A, Demissie D, Goudsmit J, Brouwer M, Kuiken C L, Pollakis G, Schuitemaker H, Fontanet A L, Rinke de Wit T F. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS. 1999;13:1305–1311. - PubMed
-
- Addo M M, Altfeld M, Rosenberg E S, Eldridge R L, Philips M N, Habeeb K, Khatri A, Brander C, Robbins G K, Mazzara G P, Goulder P J, Walker B D the HIV Controller Study Collaboration. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci USA. 2001;98:1781–1786. - PMC - PubMed
-
- Addo M M, Altfeld M, Rosenberg E S, Poon S H, Habeeb K, Philips M N, Brander C, Trocha A, Goulder P J R, Walker B D. Analysis of cytotoxic T-lymphocyte (CTL) responses against the regulatory HIV-1 proteins Rev and Tat in HIV-1-infected individuals and identification of novel CTL epitopes. Durban, South Africa: XIII International Conference on AIDS; 2000.
-
- Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothe B R, Vogel T U, Dunphy E, Liebl M E, Emerson C, Wilson N, Kunstman K J, Wang X, Allison D B, Hughes A L, Desrosiers R C, Altman J D, Wolinsky S M, Sette A, Watkins D I. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. - PubMed
-
- Altfeld M, Rosenberg E S. The role of CD4+ T helper cells in the cytotoxic T lymphocyte response to HIV-1. Curr Opin Immunol. 2000;12:375–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

