Increasing the rate of chromatin remodeling and gene activation--a novel role for the histone acetyltransferase Gcn5
- PMID: 11532958
- PMCID: PMC125614
- DOI: 10.1093/emboj/20.17.4944
Increasing the rate of chromatin remodeling and gene activation--a novel role for the histone acetyltransferase Gcn5
Abstract
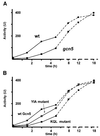
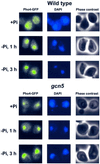
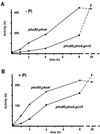
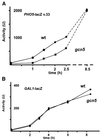
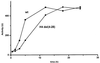
Histone acetyltransferases (HATs) such as Gcn5 play a role in transcriptional activation. However, the majority of constitutive genes show no requirement for GCN5, and even regulated genes, such as the yeast PHO5 gene, do not seem to be affected significantly by its absence under normal activation conditions. Here we show that even though the steady-state level of activated PHO5 transcription is not affected by deletion of GCN5, the rate of activation following phosphate starvation is significantly decreased. This delay in transcriptional activation is specifically due to slow chromatin remodeling of the PHO5 promoter, whereas the transmission of the phosphate starvation signal to the PHO5 promoter progresses at a normal rate. Chromatin remodeling is equally delayed in a galactose-inducible PHO5 promoter variant in which the Pho4 binding sites have been replaced by Gal4 binding sites. By contrast, activation of the GAL1 gene by galactose addition occurs with normal kinetics. Lack of the histone H4 N-termini leads to a similar delay in activation of the PHO5 promoter. These results indicate that one important contribution of HATs is to increase the rate of gene induction by accelerating chromatin remodeling, rather than to affect the final steady-state expression levels.
Figures
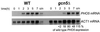

Similar articles
-
Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation.EMBO J. 2004 Jul 7;23(13):2597-607. doi: 10.1038/sj.emboj.7600230. Epub 2004 Jun 3. EMBO J. 2004. PMID: 15175650 Free PMC article.
-
The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5.EMBO J. 1999 Jun 1;18(11):3101-6. doi: 10.1093/emboj/18.11.3101. EMBO J. 1999. PMID: 10357821 Free PMC article.
-
The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene.J Mol Biol. 2002 Feb 22;316(3):489-99. doi: 10.1006/jmbi.2001.5383. J Mol Biol. 2002. PMID: 11866513
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
Histone acetylation and deacetylation in yeast.Nat Rev Mol Cell Biol. 2003 Apr;4(4):276-84. doi: 10.1038/nrm1075. Nat Rev Mol Cell Biol. 2003. PMID: 12671650 Review.
Cited by
-
Antisense non-coding transcription represses the PHO5 model gene at the level of promoter chromatin structure.PLoS Genet. 2022 Oct 10;18(10):e1010432. doi: 10.1371/journal.pgen.1010432. eCollection 2022 Oct. PLoS Genet. 2022. PMID: 36215302 Free PMC article.
-
Corepressor-directed preacetylation of histone H3 in promoter chromatin primes rapid transcriptional switching of cell-type-specific genes in yeast.Mol Cell Biol. 2010 Jul;30(13):3342-56. doi: 10.1128/MCB.01450-09. Epub 2010 May 3. Mol Cell Biol. 2010. PMID: 20439496 Free PMC article.
-
Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation.EMBO J. 2004 Jul 7;23(13):2597-607. doi: 10.1038/sj.emboj.7600230. Epub 2004 Jun 3. EMBO J. 2004. PMID: 15175650 Free PMC article.
-
Biochemical profiling of histone binding selectivity of the yeast bromodomain family.PLoS One. 2010 Jan 26;5(1):e8903. doi: 10.1371/journal.pone.0008903. PLoS One. 2010. PMID: 20126658 Free PMC article.
-
Systematic dissection of roles for chromatin regulators in a yeast stress response.PLoS Biol. 2012;10(7):e1001369. doi: 10.1371/journal.pbio.1001369. Epub 2012 Jul 31. PLoS Biol. 2012. PMID: 22912562 Free PMC article.
References
-
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

