Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application
- PMID: 11532216
- PMCID: PMC55329
- DOI: 10.1186/gb-2001-2-8-research0032
Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application
Abstract
Background: A model-based analysis of oligonucleotide expression arrays we developed previously uses a probe-sensitivity index to capture the response characteristic of a specific probe pair and calculates model-based expression indexes (MBEI). MBEI has standard error attached to it as a measure of accuracy. Here we investigate the stability of the probe-sensitivity index across different tissue types, the reproducibility of results in replicate experiments, and the use of MBEI in perfect match (PM)-only arrays.
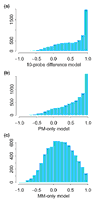
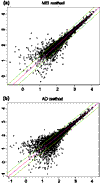
Results: Probe-sensitivity indexes are stable across tissue types. The target gene's presence in many arrays of an array set allows the probe-sensitivity index to be estimated accurately. We extended the model to obtain expression values for PM-only arrays, and found that the 20-probe PM-only model is comparable to the 10-probe PM/MM difference model, in terms of the expression correlations with the original 20-probe PM/MM difference model. MBEI method is able to extend the reliable detection limit of expression to a lower mRNA concentration. The standard errors of MBEI can be used to construct confidence intervals of fold changes, and the lower confidence bound of fold change is a better ranking statistic for filtering genes. We can assign reliability indexes for genes in a specific cluster of interest in hierarchical clustering by resampling clustering trees. A software dChip implementing many of these analysis methods is made available.
Conclusions: The model-based approach reduces the variability of low expression estimates, and provides a natural method of calculating expression values for PM-only arrays. The standard errors attached to expression values can be used to assess the reliability of downstream analysis.
Figures




Similar articles
-
Theoretical and experimental comparisons of gene expression indexes for oligonucleotide arrays.Bioinformatics. 2002 Nov;18(11):1470-6. doi: 10.1093/bioinformatics/18.11.1470. Bioinformatics. 2002. PMID: 12424118
-
Exploration, normalization, and summaries of high density oligonucleotide array probe level data.Biostatistics. 2003 Apr;4(2):249-64. doi: 10.1093/biostatistics/4.2.249. Biostatistics. 2003. PMID: 12925520
-
Statistical analysis of high-density oligonucleotide arrays: a multiplicative noise model.Bioinformatics. 2002 Dec;18(12):1633-40. doi: 10.1093/bioinformatics/18.12.1633. Bioinformatics. 2002. PMID: 12490448
-
Fundamentals of DNA hybridization arrays for gene expression analysis.Biotechniques. 2000 Nov;29(5):1042-6, 1048-55. doi: 10.2144/00295rv01. Biotechniques. 2000. PMID: 11084867 Review.
-
Post-analysis follow-up and validation of microarray experiments.Nat Genet. 2002 Dec;32 Suppl:509-14. doi: 10.1038/ng1034. Nat Genet. 2002. PMID: 12454646 Review.
Cited by
-
Analysis of tanshinone IIA induced cellular apoptosis in leukemia cells by genome-wide expression profiling.BMC Complement Altern Med. 2012 Jan 16;12:5. doi: 10.1186/1472-6882-12-5. BMC Complement Altern Med. 2012. PMID: 22248096 Free PMC article.
-
Genome-wide prediction and analysis of human tissue-selective genes using microarray expression data.BMC Med Genomics. 2013;6 Suppl 1(Suppl 1):S10. doi: 10.1186/1755-8794-6-S1-S10. Epub 2013 Jan 23. BMC Med Genomics. 2013. PMID: 23369200 Free PMC article.
-
Accelerated progression of chronic lymphocytic leukemia in Eμ-TCL1 mice expressing catalytically inactive RAG1.Blood. 2013 May 9;121(19):3855-66, S1-16. doi: 10.1182/blood-2012-08-446732. Epub 2013 Mar 15. Blood. 2013. PMID: 23502221 Free PMC article.
-
Preservation of ranking order in the expression of human Housekeeping genes.PLoS One. 2011;6(12):e29314. doi: 10.1371/journal.pone.0029314. Epub 2011 Dec 22. PLoS One. 2011. PMID: 22216246 Free PMC article.
-
Gene expression profiling of microglia infected by a highly neurovirulent murine leukemia virus: implications for neuropathogenesis.Retrovirology. 2006 May 12;3:26. doi: 10.1186/1742-4690-3-26. Retrovirology. 2006. PMID: 16696860 Free PMC article.
References
-
- Wodicka L, Dong H, Mittmann M, Ho M, Lockhart D. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. - PubMed
-
- Wallace D. The Behrens-Fisher and Fieller-Creasy problems. In Lecture Notes in Statistics 1, RAFisher: An Appreciation Edited by Fienberg SE, Hinkley DV Springer-Verlag. 1988. pp. 119–147.
-
- Cox DR, Hinkley DV. Theoretical Statistics London: Chapman and Hall, 1974.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

