Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons
- PMID: 11528000
- PMCID: PMC99031
- DOI: 10.1128/MMBR.65.3.353-370.2001
Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons
Abstract
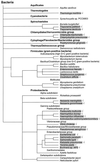
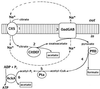
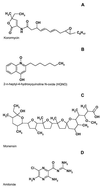
Analysis of the bacterial genome sequences shows that many human and animal pathogens encode primary membrane Na+ pumps, Na+-transporting dicarboxylate decarboxylases or Na+ translocating NADH:ubiquinone oxidoreductase, and a number of Na+ -dependent permeases. This indicates that these bacteria can utilize Na+ as a coupling ion instead of or in addition to the H+ cycle. This capability to use a Na+ cycle might be an important virulence factor for such pathogens as Vibrio cholerae, Neisseria meningitidis, Salmonella enterica serovar Typhi, and Yersinia pestis. In Treponema pallidum, Chlamydia trachomatis, and Chlamydia pneumoniae, the Na+ gradient may well be the only energy source for secondary transport. A survey of preliminary genome sequences of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and Treponema denticola indicates that these oral pathogens also rely on the Na+ cycle for at least part of their energy metabolism. The possible roles of the Na+ cycling in the energy metabolism and pathogenicity of these organisms are reviewed. The recent discovery of an effective natural antibiotic, korormicin, targeted against the Na+ -translocating NADH:ubiquinone oxidoreductase, suggests a potential use of Na+ pumps as drug targets and/or vaccine candidates. The antimicrobial potential of other inhibitors of the Na+ cycle, such as monensin, Li+ and Ag+ ions, and amiloride derivatives, is discussed.
Figures

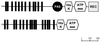
Similar articles
-
Antibiotic Korormicin A Kills Bacteria by Producing Reactive Oxygen Species.J Bacteriol. 2019 May 8;201(11):e00718-18. doi: 10.1128/JB.00718-18. Print 2019 Jun 1. J Bacteriol. 2019. PMID: 30858300 Free PMC article.
-
Salt in the wound: a possible role of Na+ gradient in chlamydial infection.J Mol Microbiol Biotechnol. 2004;8(1):1-6. doi: 10.1159/000082075. J Mol Microbiol Biotechnol. 2004. PMID: 15741735 Review.
-
Recent progress in the Na(+)-translocating NADH-quinone reductase from the marine Vibrio alginolyticus.Biochim Biophys Acta. 2001 May 1;1505(1):37-44. doi: 10.1016/s0005-2728(00)00275-9. Biochim Biophys Acta. 2001. PMID: 11248187 Review.
-
Inhibitor studies of a new antibiotic, korormicin, 2-n-heptyl-4-hydroxyquinoline N-oxide and Ag+ toward the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus.Biol Pharm Bull. 1999 Oct;22(10):1064-7. doi: 10.1248/bpb.22.1064. Biol Pharm Bull. 1999. PMID: 10549856
-
NqrM (DUF539) Protein Is Required for Maturation of Bacterial Na+-Translocating NADH:Quinone Oxidoreductase.J Bacteriol. 2015 Dec 7;198(4):655-63. doi: 10.1128/JB.00757-15. J Bacteriol. 2015. PMID: 26644436 Free PMC article.
Cited by
-
Effect of respiratory inhibitors and quinone analogues on the aerobic electron transport system of Eikenella corrodens.Sci Rep. 2021 Apr 26;11(1):8987. doi: 10.1038/s41598-021-88388-0. Sci Rep. 2021. PMID: 33903681 Free PMC article.
-
Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools.Sci Rep. 2013;3:1748. doi: 10.1038/srep01748. Sci Rep. 2013. PMID: 23624511 Free PMC article.
-
Mutation of two key aspartate residues alters stoichiometry of the NhaB Na+/H+ exchanger from Klebsiella pneumoniae.Sci Rep. 2019 Oct 28;9(1):15390. doi: 10.1038/s41598-019-51887-2. Sci Rep. 2019. PMID: 31659210 Free PMC article.
-
Bacterial signal transduction network in a genomic perspective.Environ Microbiol. 2004 Jun;6(6):552-67. doi: 10.1111/j.1462-2920.2004.00633.x. Environ Microbiol. 2004. PMID: 15142243 Free PMC article. Review.
-
Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay.Microb Ecol. 2004 Jan;47(1):41-7. doi: 10.1007/s00248-003-1032-9. Microb Ecol. 2004. PMID: 15259268
References
-
- Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. - PubMed
-
- Atsumi T, Maekawa Y, Tokuda H, Imae Y. Amiloride at pH 7.0 inhibits the Na+-driven flagellar motors of Vibrio alginolyticus but allows cell growth. FEBS Lett. 1992;314:114–116. - PubMed
-
- Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

