Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism
- PMID: 11507195
- PMCID: PMC115095
- DOI: 10.1128/jvi.75.18.8498-8506.2001
Human immunodeficiency virus type 1 IIIB selected for replication in vivo exhibits increased envelope glycoproteins in virions without alteration in coreceptor usage: separation of in vivo replication from macrophage tropism
Abstract
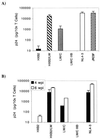
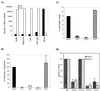
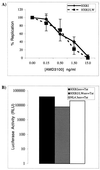
Analysis of viral replication and pathogenicity after in vivo selection of human immunodeficiency virus type 1 (HIV-1) attenuated in vitro will help to define the functions involved in replication and pathogenesis in vivo. Using the SCID-hu Thy/Liv mouse and human fetal thymus organ culture as in vivo models, we previously defined HIV-1 env determinants (HXB2/LW) which were reverted for replication in vivo (L. Su et al., Virology 227:46-52, 1997). In this study, we examined the replication of four highly related HIV-1 clones directly derived from Lai/IIIB or after selection in vivo to investigate the envelope gp120 determinants associated with replication in macrophages and in the thymus models in vivo. The LW/C clone derived from the IIIB-infected laboratory worker and HXB2/LW both efficiently infected monocyte-derived macrophages (MDM) and the human thymus. Although the laboratory worker (LW) isolates showed altered tropism from IIIB, they still predominantly used CXCR4 as coreceptors for infecting peripheral blood mononuclear cells, macrophages, and the thymus. Interestingly, a single amino acid mutation in the V3 loop associated with resistance to neutralizing antibodies was also essential for the replication activity of the LW virus in the thymus models but not for its activity in infecting MDM. The LW virions were equally sensitive to a CXCR4 antagonist. We further demonstrated that the LW HIV-1 isolate selected in vivo produced more infectious viral particles that contained higher levels of the Env protein gp120. Thus, selection of the laboratory-attenuated Lai/IIIB isolate in vivo leads to altered tropism but not coreceptor usage of the virus. The acquired replication activity in vivo is correlated with an early A-to-T mutation in the V3 loop and increased virion association of HIV-1 Env gp120, but it is genetically separable from the acquired replication activity in macrophages.
Figures

Similar articles
-
Identification of HIV-1 determinants for replication in vivo.Virology. 1997 Jan 6;227(1):45-52. doi: 10.1006/viro.1996.8338. Virology. 1997. PMID: 9007057
-
Separation of human immunodeficiency virus type 1 replication from nef-mediated pathogenesis in the human thymus.J Virol. 2001 Apr;75(8):3916-24. doi: 10.1128/JVI.75.8.3916-3924.2001. J Virol. 2001. PMID: 11264380 Free PMC article.
-
Role of naturally occurring basic amino acid substitutions in the human immunodeficiency virus type 1 subtype E envelope V3 loop on viral coreceptor usage and cell tropism.J Virol. 1999 Jul;73(7):5520-6. doi: 10.1128/JVI.73.7.5520-5526.1999. J Virol. 1999. PMID: 10364300 Free PMC article.
-
HIV-1 receptors and cell tropism.Br Med Bull. 2001;58:43-59. doi: 10.1093/bmb/58.1.43. Br Med Bull. 2001. PMID: 11714623 Review.
-
HIV-1 envelope determinants for cell tropism and chemokine receptor use.Mol Membr Biol. 1999 Jan-Mar;16(1):57-65. doi: 10.1080/096876899294760. Mol Membr Biol. 1999. PMID: 10332738 Review.
Cited by
-
The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 Env.J Virol. 2009 Nov;83(22):11715-25. doi: 10.1128/JVI.00649-09. Epub 2009 Sep 2. J Virol. 2009. PMID: 19726524 Free PMC article.
-
Adoption of an "open" envelope conformation facilitating CD4 binding and structural remodeling precedes coreceptor switch in R5 SHIV-infected macaques.PLoS One. 2011;6(7):e21350. doi: 10.1371/journal.pone.0021350. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760891 Free PMC article.
-
Conserved changes in envelope function during human immunodeficiency virus type 1 coreceptor switching.J Virol. 2007 Aug;81(15):8165-79. doi: 10.1128/JVI.02792-06. Epub 2007 May 16. J Virol. 2007. PMID: 17507486 Free PMC article.
-
HIV-1 replication and pathogenesis in the human thymus.Curr HIV Res. 2003 Jul;1(3):275-85. doi: 10.2174/1570162033485258. Curr HIV Res. 2003. PMID: 15046252 Free PMC article. Review.
-
Fusion-induced apoptosis contributes to thymocyte depletion by a pathogenic human immunodeficiency virus type 1 envelope in the human thymus.J Virol. 2006 Nov;80(22):11019-30. doi: 10.1128/JVI.01382-06. Epub 2006 Sep 6. J Virol. 2006. PMID: 16956934 Free PMC article.
References
-
- Aldrovandi G M, Feuer G, Gao L, Jamieson B, Kristeva M, Chen I S, Zack J A. The SCID-hu mouse as a model for HIV-1 infection. Nature. 1993;363:732–736. - PubMed
-
- Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

