Kinetic analysis of the steps of the polyomavirus lytic cycle
- PMID: 11507182
- PMCID: PMC115082
- DOI: 10.1128/jvi.75.18.8368-8379.2001
Kinetic analysis of the steps of the polyomavirus lytic cycle
Abstract
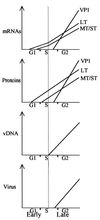
Kinetic studies of the accumulation of early and late transcripts, early and late proteins, genomes, and live virus, during the lytic cycle of murine polyomavirus wild-type A2, were carried out in synchronized NIH 3T3 cells released from G(0) by the addition of serum after infection. This first-time simultaneous analysis of all parameters of the virus life cycle led to new insights concerning the transcriptional control at the early-to-late transition. During the early phase, early transcripts were synthesized at very low levels, detectable only by reverse transcription-PCR, from 6 h postinfection (hpi). Large T protein could be detected by 8 hpi (while infected cells were in the G(1) phase). The level of expression of the middle T and small T proteins was lower than that of large T at all times, due, at least in part, to a splicing preference for the large-T 5' splice site at nucleotide 411. A large increase in the level of both early and late transcripts coincided closely with the detection in mid-S phase of viral genome amplification. Thereafter, both classes of transcripts continued to further accumulate up to the end of the experiments (48 hpi). In addition, during the late phase, "giant" multigenomic transcripts were synthesized from the early as well as the late promoter. Thus, a major type of transcriptional control appears to be applied similarly to the transcription of both early and late genes. This view differs from that in the literature, which highlights the enhancement of late transcription and the repression of early transcription. However, despite this parallel transcriptional control, additional regulations are applied which result in higher levels of late compared to early transcripts, as previously described. In the accompanying article, a key role for middle T and/or small T in this late-phase enhancement of early and late transcription is demonstrated (16). Other novel findings, e.g., the synthesis of a very abundant short early promoter proximal RNA, are also described.
Figures







Similar articles
-
Role of middle T-small T in the lytic cycle of polyomavirus: control of the early-to-late transcriptional switch and viral DNA replication.J Virol. 2001 Sep;75(18):8380-9. doi: 10.1128/jvi.75.18.8380-8389.2001. J Virol. 2001. PMID: 11507183 Free PMC article.
-
Small and middle T antigens contribute to lytic and abortive polyomavirus infection.J Virol. 1985 Feb;53(2):579-86. doi: 10.1128/JVI.53.2.579-586.1985. J Virol. 1985. PMID: 2578576 Free PMC article.
-
Enhancer-mediated role for polyomavirus middle T/small T in DNA replication.J Virol. 1995 Jan;69(1):326-33. doi: 10.1128/JVI.69.1.326-333.1995. J Virol. 1995. PMID: 7983726 Free PMC article.
-
Role of mouse polyomavirus late region in the control of viral DNA replication: a review.Biochimie. 1995;77(10):780-6. doi: 10.1016/0300-9084(96)88196-x. Biochimie. 1995. PMID: 8824775 Review.
-
How a small DNA virus uses dsRNA but not RNAi to regulate its life cycle.Cold Spring Harb Symp Quant Biol. 2006;71:293-9. doi: 10.1101/sqb.2006.71.017. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381309 Review.
Cited by
-
Stimulation of DNA replication from the polyomavirus origin by PCAF and GCN5 acetyltransferases: acetylation of large T antigen.Mol Cell Biol. 2002 Nov;22(22):7907-18. doi: 10.1128/MCB.22.22.7907-7918.2002. Mol Cell Biol. 2002. PMID: 12391158 Free PMC article.
-
Role of middle T-small T in the lytic cycle of polyomavirus: control of the early-to-late transcriptional switch and viral DNA replication.J Virol. 2001 Sep;75(18):8380-9. doi: 10.1128/jvi.75.18.8380-8389.2001. J Virol. 2001. PMID: 11507183 Free PMC article.
-
Inhibition of polyomavirus ori-dependent DNA replication by mSin3B.J Virol. 2002 Dec;76(23):11809-18. doi: 10.1128/jvi.76.23.11809-11818.2002. J Virol. 2002. PMID: 12414923 Free PMC article.
-
Immune sensing of mouse polyomavirus DNA by p204 and cGAS DNA sensors.FEBS J. 2021 Oct;288(20):5964-5985. doi: 10.1111/febs.15962. Epub 2021 May 26. FEBS J. 2021. PMID: 33969628 Free PMC article.
-
Induction of the human papillomavirus type 31 late promoter requires differentiation but not DNA amplification.J Virol. 2005 Apr;79(8):4918-26. doi: 10.1128/JVI.79.8.4918-4926.2005. J Virol. 2005. PMID: 15795277 Free PMC article.
References
-
- Acheson N H. Lytic cycle of SV40 and polyoma virus. In: Tooze J, editor. DNA tumor virus. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1981. pp. 125–204.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

