An Eulerian path approach to DNA fragment assembly
- PMID: 11504945
- PMCID: PMC55524
- DOI: 10.1073/pnas.171285098
An Eulerian path approach to DNA fragment assembly
Abstract
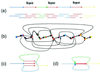
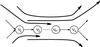
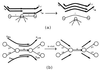
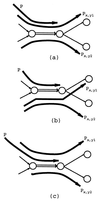
For the last 20 years, fragment assembly in DNA sequencing followed the "overlap-layout-consensus" paradigm that is used in all currently available assembly tools. Although this approach proved useful in assembling clones, it faces difficulties in genomic shotgun assembly. We abandon the classical "overlap-layout-consensus" approach in favor of a new euler algorithm that, for the first time, resolves the 20-year-old "repeat problem" in fragment assembly. Our main result is the reduction of the fragment assembly to a variation of the classical Eulerian path problem that allows one to generate accurate solutions of large-scale sequencing problems. euler, in contrast to the celera assembler, does not mask such repeats but uses them instead as a powerful fragment assembly tool.
Figures

Similar articles
-
Fragment assembly with double-barreled data.Bioinformatics. 2001;17 Suppl 1:S225-33. doi: 10.1093/bioinformatics/17.suppl_1.s225. Bioinformatics. 2001. PMID: 11473013
-
Correcting base-assignment errors in repeat regions of shotgun assembly.IEEE/ACM Trans Comput Biol Bioinform. 2007 Jan-Mar;4(1):54-64. doi: 10.1109/TCBB.2007.1005. IEEE/ACM Trans Comput Biol Bioinform. 2007. PMID: 17277413
-
Short read fragment assembly of bacterial genomes.Genome Res. 2008 Feb;18(2):324-30. doi: 10.1101/gr.7088808. Epub 2007 Dec 14. Genome Res. 2008. PMID: 18083777 Free PMC article.
-
Assembly algorithms for next-generation sequencing data.Genomics. 2010 Jun;95(6):315-27. doi: 10.1016/j.ygeno.2010.03.001. Epub 2010 Mar 6. Genomics. 2010. PMID: 20211242 Free PMC article. Review.
-
Sequence assembly using next generation sequencing data--challenges and solutions.Sci China Life Sci. 2014 Nov;57(11):1140-8. doi: 10.1007/s11427-014-4752-9. Epub 2014 Oct 17. Sci China Life Sci. 2014. PMID: 25326069 Review.
Cited by
-
Space-time Trade-offs for the LCP Array of Wheeler DFAs.Int Symp String Process Inf Retr. 2023 Sep;14240:143-156. doi: 10.1007/978-3-031-43980-3_12. Epub 2023 Sep 20. Int Symp String Process Inf Retr. 2023. PMID: 39108943 Free PMC article.
-
Unlocking plant genetics with telomere-to-telomere genome assemblies.Nat Genet. 2024 Sep;56(9):1788-1799. doi: 10.1038/s41588-024-01830-7. Epub 2024 Jul 24. Nat Genet. 2024. PMID: 39048791 Review.
-
Unitig-centered pan-genome machine learning approach for predicting antibiotic resistance and discovering novel resistance genes in bacterial strains.Comput Struct Biotechnol J. 2024 Apr 16;23:1864-1876. doi: 10.1016/j.csbj.2024.04.035. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38707536 Free PMC article.
-
Revisiting the complexity of and algorithms for the graph traversal edit distance and its variants.Algorithms Mol Biol. 2024 Apr 29;19(1):17. doi: 10.1186/s13015-024-00262-6. Algorithms Mol Biol. 2024. PMID: 38679703 Free PMC article.
-
Genome assembly in the telomere-to-telomere era.Nat Rev Genet. 2024 Sep;25(9):658-670. doi: 10.1038/s41576-024-00718-w. Epub 2024 Apr 22. Nat Rev Genet. 2024. PMID: 38649458 Review.
References
-
- Green P. Documentation for phrap. 1994. (http://www.genome.washington.edu/UWGC/analysis-tools/phrap.htm).
-
- Sutton G, White O, Adams M, Kerlavage A. Genome Sci Technol. 1995;1:9–19.
-
- Kececioglu J, Myers E. Algorithmica. 1995;13:7–51.
-
- Myers E M. J Comput Biol. 1995;2:275–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

