A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface
- PMID: 11481477
- PMCID: PMC55380
- DOI: 10.1073/pnas.161282998
A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface
Abstract
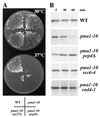
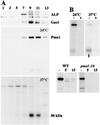
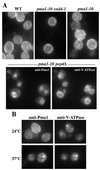
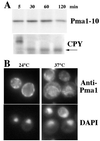
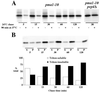
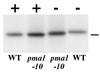
Pma1 is a plasma membrane H(+)-ATPase whose activity at the cell surface is essential for cell viability. In this paper we describe a temperature-sensitive pma1 allele, pma1-10 (with two point mutations in the first cytoplasmic loop of Pma1), in which the newly synthesized mutant protein fails to remain stable at the cell surface at 37 degrees C. Instead, Pma1-10 appears to undergo internalization for vacuolar degradation in a manner dependent on End4, Vps27, Doa4, and Pep4. By contrast with wild-type Pma1, mutant Pma1-10 is hypophosphorylated and fails to associate with a Triton-insoluble fraction at 37 degrees C, suggesting failure to enter lipid rafts. Kinetic analysis reveals that, at the permissive temperature, newly synthesized Pma1-10 acquires Triton-insolubility before becoming stabilized. We suggest that phosphorylation and lipid raft association may play important roles in maintaining protein stability at the plasma membrane.
Figures
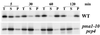
Similar articles
-
An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast.Mol Biol Cell. 2000 Feb;11(2):579-92. doi: 10.1091/mbc.11.2.579. Mol Biol Cell. 2000. PMID: 10679016 Free PMC article.
-
Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole.J Cell Biol. 1995 Jan;128(1-2):39-49. doi: 10.1083/jcb.128.1.39. J Cell Biol. 1995. PMID: 7822420 Free PMC article.
-
Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast.Mol Biol Cell. 2004 May;15(5):2401-9. doi: 10.1091/mbc.e03-10-0727. Epub 2004 Mar 12. Mol Biol Cell. 2004. PMID: 15020711 Free PMC article.
-
Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast.Biochimie. 2007 Feb;89(2):249-54. doi: 10.1016/j.biochi.2006.07.020. Epub 2006 Aug 15. Biochimie. 2007. PMID: 16938383 Review.
-
Structure and function of the yeast vacuolar membrane proton ATPase.J Bioenerg Biomembr. 1989 Oct;21(5):589-603. doi: 10.1007/BF00808115. J Bioenerg Biomembr. 1989. PMID: 2531738 Review.
Cited by
-
Quality control for unfolded proteins at the plasma membrane.J Cell Biol. 2010 Nov 1;191(3):553-70. doi: 10.1083/jcb.201006012. Epub 2010 Oct 25. J Cell Biol. 2010. PMID: 20974815 Free PMC article.
-
The 3'-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein.RNA. 2008 Jul;14(7):1352-65. doi: 10.1261/rna.867208. Epub 2008 May 20. RNA. 2008. PMID: 18492794 Free PMC article.
-
The Essential Neo1 Protein from Budding Yeast Plays a Role in Establishing Aminophospholipid Asymmetry of the Plasma Membrane.J Biol Chem. 2016 Jul 22;291(30):15727-39. doi: 10.1074/jbc.M115.686253. Epub 2016 May 26. J Biol Chem. 2016. PMID: 27235400 Free PMC article.
-
Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans.Mol Biol Cell. 2004 Dec;15(12):5528-37. doi: 10.1091/mbc.e04-08-0666. Epub 2004 Sep 15. Mol Biol Cell. 2004. PMID: 15371534 Free PMC article.
-
Protein phosphatase type 1 regulates ion homeostasis in Saccharomyces cerevisiae.Genetics. 2002 Apr;160(4):1423-37. doi: 10.1093/genetics/160.4.1423. Genetics. 2002. PMID: 11973298 Free PMC article.
References
-
- Serrano R, Kielland-Brandt M C, Fink G R. Nature (London) 1986;319:689–693. - PubMed
-
- Auer M, Scarborough G A, Kuhlbrandt W. Nature (London) 1998;392:840–843. - PubMed
-
- Morsomme P, Slayman C W, Goffeau A. Biochim Biophys Acta. 2000;1469:133–157. - PubMed
-
- DeWitt N D, Tourinho dos Santos C F, Allen K E, Slayman C W. J Biol Chem. 1998;273:21744–21751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

