Conformational switch and role of phosphorylation in PAK activation
- PMID: 11438672
- PMCID: PMC87242
- DOI: 10.1128/MCB.21.15.5179-5189.2001
Conformational switch and role of phosphorylation in PAK activation
Abstract
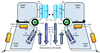
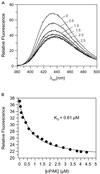
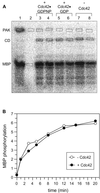
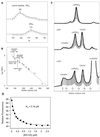
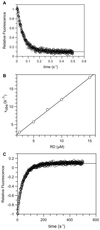
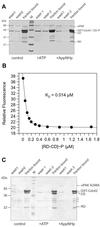
p21-activated protein kinases (PAKs) are involved in signal transduction processes initiating a variety of biological responses. They become activated by interaction with Rho-type small GTP-binding proteins Rac and Cdc42 in the GTP-bound conformation, thereby relieving the inhibition of the regulatory domain (RD) on the catalytic domain (CD). Here we report on the mechanism of activation and show that proteolytic digestion of PAK produces a heterodimeric RD-CD complex consisting of a regulatory fragment (residues 57 to 200) and a catalytic fragment (residues 201 to 491), which is active in the absence of Cdc42. Cdc42-GppNHp binds with low affinity (K(d) 0.6 microM) to intact kinase, whereas the affinity to the isolated regulatory fragment is much higher (K(d) 18 nM), suggesting that the difference in binding energy is used for the conformational change leading to activation. The full-length kinase, the isolated RD, and surprisingly also their complexes with Cdc42 behave as dimers on a gel filtration column. Cdc42-GppNHp interaction with the RD-CD complex is also of low affinity and does not dissociate the RD from the CD. After autophosphorylation of the kinase domain, Cdc42 binds with high (14 nM) affinity and dissociates the RD-CD complex. Assuming that the RD-CD complex mimics the interaction in native PAK, this indicates that the small G protein may not simply release the RD from the CD. It acts in a more subtle allosteric control mechanism to induce autophosphorylation, which in turn induces the release of the RD and thus full activation.
Figures



Similar articles
-
Delineation of the Cdc42/Rac-binding domain of p21-activated kinase.Biochemistry. 1998 May 26;37(21):7885-91. doi: 10.1021/bi980140+. Biochemistry. 1998. PMID: 9601050
-
A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases.J Mol Biol. 2006 Aug 11;361(2):312-26. doi: 10.1016/j.jmb.2006.06.017. Epub 2006 Jun 27. J Mol Biol. 2006. PMID: 16837009
-
Oncogenic Dbl, Cdc42, and p21-activated kinase form a ternary signaling intermediate through the minimum interactive domains.Biochemistry. 2004 Nov 23;43(46):14584-93. doi: 10.1021/bi048574u. Biochemistry. 2004. PMID: 15544329
-
Involvement of Rac/Cdc42/PAK pathway in cytoskeletal rearrangements.Acta Biochim Pol. 2009;56(2):225-34. Epub 2009 Jun 10. Acta Biochim Pol. 2009. PMID: 19513348 Review.
-
The genetics of Pak.J Cell Sci. 2004 Sep 1;117(Pt 19):4343-54. doi: 10.1242/jcs.01392. J Cell Sci. 2004. PMID: 15331659 Review.
Cited by
-
p21-Activated Kinase: Role in Gastrointestinal Cancer and Beyond.Cancers (Basel). 2022 Sep 28;14(19):4736. doi: 10.3390/cancers14194736. Cancers (Basel). 2022. PMID: 36230657 Free PMC article. Review.
-
The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK.EMBO J. 2004 Jul 21;23(14):2777-88. doi: 10.1038/sj.emboj.7600291. Epub 2004 Jul 1. EMBO J. 2004. PMID: 15229651 Free PMC article.
-
Roles of P21-activated kinases and associated proteins in epithelial wound healing.Int Rev Cell Mol Biol. 2008;267:253-98. doi: 10.1016/S1937-6448(08)00606-0. Int Rev Cell Mol Biol. 2008. PMID: 18544501 Free PMC article. Review.
-
Validation of the p21-activated kinases as targets for inhibition in neurofibromatosis type 2.Cancer Res. 2008 Oct 1;68(19):7932-7. doi: 10.1158/0008-5472.CAN-08-0866. Cancer Res. 2008. PMID: 18829550 Free PMC article.
-
PAK signalling during the development and progression of cancer.Nat Rev Cancer. 2014 Jan;14(1):13-25. doi: 10.1038/nrc3645. Nat Rev Cancer. 2014. PMID: 24505617 Free PMC article. Review.
References
-
- Abdul-Manan N, Aghazadeh B, Liu G A, Majumdar A, Ouerfelli O, Siminovitch K A, Rosen M K. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–383. - PubMed
-
- Bagrodia S, Cerione R A. PAK to the future. Trends Cell Biol. 1999;9:350–355. - PubMed
-
- Benner G E, Dennis P B, Masaracchia R A. Activation of an s6/h4 kinase (Pak 65) from human placenta by intramolecular and intermolecular autophosphorylation. J Biol Chem. 1995;270:21121–21128. - PubMed
-
- Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. - PubMed
-
- Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
