PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator
- PMID: 11432836
- PMCID: PMC125516
- DOI: 10.1093/emboj/20.13.3495
PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator
Abstract
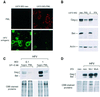
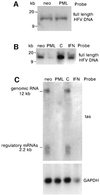
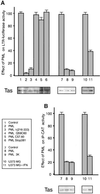
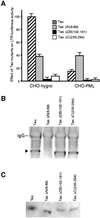
The promyelocytic leukaemia (PML) protein localizes in the nucleus both in the nucleoplasm and in matrix-associated multiprotein complexes known as nuclear bodies (NBs). The number and the intensity of PML NBs increase in response to interferon (IFN). Overexpression of PML affects the replication of vesicular stomatitis virus and influenza virus. However, PML has a less powerful antiviral activity against these viruses than the IFN mediator MxA. Here, we show that overexpression of PML, but not that of Mx1 or MxA, leads to a drastic decrease of a complex retrovirus, the human foamy virus (HFV), gene expression. PML represses HFV transcription by complexing the HFV transactivator, Tas, preventing its direct binding to viral DNA. This physical interaction requires the N-terminal region of Tas and the RING finger of PML, but does not necessitate PML localization in NBs. Finally, we show that IFN treatment inhibits HFV replication in wild-type but not in PML-/- cells. These findings point to a role for PML in transcriptional repression and suggest that PML could play a key role in mediating an IFN-induced antiviral state against a complex retrovirus.
Figures


Similar articles
-
Role and fate of PML nuclear bodies in response to interferon and viral infections.Oncogene. 2001 Oct 29;20(49):7274-86. doi: 10.1038/sj.onc.1204854. Oncogene. 2001. PMID: 11704856 Review.
-
Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies.Exp Cell Res. 2001 Dec 10;271(2):286-95. doi: 10.1006/excr.2001.5380. Exp Cell Res. 2001. PMID: 11716541
-
Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection.Virology. 2000 Aug 15;274(1):39-55. doi: 10.1006/viro.2000.0448. Virology. 2000. PMID: 10936087
-
The promyelocytic leukemia protein does not mediate foamy virus latency in vitro.J Virol. 2003 Feb;77(3):2207-13. doi: 10.1128/jvi.77.3.2207-2213.2003. J Virol. 2003. PMID: 12525655 Free PMC article.
-
[Alpha interferon, antiviral proteins and their value in clinical medicine].Ann Biol Clin (Paris). 1999 Nov-Dec;57(6):659-66. Ann Biol Clin (Paris). 1999. PMID: 10572214 Review. French.
Cited by
-
Interaction of herpes simplex virus ICP0 with ND10 bodies: a sequential process of adhesion, fusion, and retention.J Virol. 2013 Sep;87(18):10244-54. doi: 10.1128/JVI.01487-13. Epub 2013 Jul 17. J Virol. 2013. PMID: 23864622 Free PMC article.
-
Virion factors that target Daxx to overcome intrinsic immunity.J Virol. 2013 Oct;87(19):10412-22. doi: 10.1128/JVI.00425-13. Epub 2013 Jul 17. J Virol. 2013. PMID: 23864634 Free PMC article. Review.
-
Leishmania infection upregulates and engages host macrophage Argonaute 1, and system-wide proteomics reveals Argonaute 1-dependent host response.Front Immunol. 2023 Nov 30;14:1287539. doi: 10.3389/fimmu.2023.1287539. eCollection 2023. Front Immunol. 2023. PMID: 38098491 Free PMC article.
-
Inhibition of spumavirus gene expression by PHF11.PLoS Pathog. 2020 Jul 17;16(7):e1008644. doi: 10.1371/journal.ppat.1008644. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32678836 Free PMC article.
-
Patterns of evolution of host proteins involved in retroviral pathogenesis.Retrovirology. 2006 Feb 7;3:11. doi: 10.1186/1742-4690-3-11. Retrovirology. 2006. PMID: 16460575 Free PMC article. Review.
References
-
- Chelbi-Alix M. and de Thé,H. (1999) Herpes virus induces proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100. Oncogene, 18, 935–941. - PubMed
-
- Chelbi-Alix M.K., Pelicano,L., Quignon,F., Koken,M.H.M., Venturini,L., Stadler,M., Pavlovic,J., Degos,L. and de Thé,H. (1995) Induction of the PML protein by interferons in normal and APL cells. Leukemia, 9, 2027–2033. - PubMed
-
- Chelbi-Alix M.K., Pelicano,L., Quignon,F., Koken,M.H.M. and de Thé,H. (1996) PML is a primary target gene of interferon and could mediate some of its biological activities. Tumor Biol., 99, 17–27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

