Essential role of cyclization sequences in flavivirus RNA replication
- PMID: 11413342
- PMCID: PMC114398
- DOI: 10.1128/JVI.75.14.6719-6728.2001
Essential role of cyclization sequences in flavivirus RNA replication
Abstract
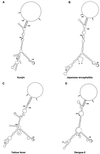
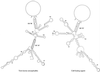
A possible role in RNA replication for interactions between conserved complementary (cyclization) sequences in the 5'- and 3'-terminal regions of Flavivirus RNA was previously suggested but never tested in vivo. Using the M-fold program for RNA secondary-structure predictions, we examined for the first time the base-pairing interactions between the covalently linked 5' genomic region (first ~160 nucleotides) and the 3' untranslated region (last ~115 nucleotides) for a range of mosquito-borne Flavivirus species. Base-pairing occurred as predicted for the previously proposed conserved cyclization sequences. In order to obtain experimental evidence of the predicted interactions, the putative cyclization sequences (5' or 3') in the replicon RNA of the mosquito-borne Kunjin virus were mutated either separately, to destroy base-pairing, or simultaneously, to restore the complementarity. None of the RNAs with separate mutations in only the 5' or only the 3' cyclization sequences was able to replicate after transfection into BHK cells, while replicon RNA with simultaneous compensatory mutations in both cyclization sequences was replication competent. This was detected by immunofluorescence for expression of the major nonstructural protein NS3 and by Northern blot analysis for amplification and accumulation of replicon RNA. We then used the M-fold program to analyze RNA secondary structure of the covalently linked 5'- and 3'-terminal regions of three tick-borne virus species and identified a previously undescribed additional pair of conserved complementary sequences in locations similar to those of the mosquito-borne species. They base-paired with DeltaG values of approximately -20 kcal, equivalent or greater in stability than those calculated for the originally proposed cyclization sequences. The results show that the base-pairing between 5' and 3' complementary sequences, rather than the nucleotide sequence per se, is essential for the replication of mosquito-borne Kunjin virus RNA and that more than one pair of cyclization sequences might be involved in the replication of the tick-borne Flavivirus species.
Figures




Similar articles
-
Functional analysis of dengue virus cyclization sequences located at the 5' and 3'UTRs.Virology. 2008 May 25;375(1):223-35. doi: 10.1016/j.virol.2008.01.014. Epub 2008 Mar 4. Virology. 2008. PMID: 18289628
-
An RNA domain within the 5' untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication.J Mol Biol. 2001 Jan 26;305(4):741-56. doi: 10.1006/jmbi.2000.4298. J Mol Biol. 2001. PMID: 11162089
-
Sequence comparison and secondary structure analysis of the 3' noncoding region of flavivirus genomes reveals multiple pseudoknots.RNA. 2001 Oct;7(10):1370-7. RNA. 2001. PMID: 11680841 Free PMC article.
-
Functions of the 3' and 5' genome RNA regions of members of the genus Flavivirus.Virus Res. 2015 Aug 3;206:108-19. doi: 10.1016/j.virusres.2015.02.006. Epub 2015 Feb 13. Virus Res. 2015. PMID: 25683510 Free PMC article. Review.
-
Genome cyclization as strategy for flavivirus RNA replication.Virus Res. 2009 Feb;139(2):230-9. doi: 10.1016/j.virusres.2008.07.016. Epub 2008 Sep 9. Virus Res. 2009. PMID: 18703097 Free PMC article. Review.
Cited by
-
Polymerase Spiral Reaction Assay for Rapid and Real Time Detection of West Nile Virus From Clinical Samples.Front Cell Infect Microbiol. 2020 Aug 27;10:426. doi: 10.3389/fcimb.2020.00426. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32984063 Free PMC article.
-
Replacement of conserved or variable sequences of the mosquito-borne dengue virus 3' UTR with homologous sequences from Modoc virus does not change infectivity for mosquitoes.J Gen Virol. 2013 Apr;94(Pt 4):783-788. doi: 10.1099/vir.0.046664-0. Epub 2012 Dec 19. J Gen Virol. 2013. PMID: 23255623 Free PMC article.
-
Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription.RNA. 2010 Jun;16(6):1226-35. doi: 10.1261/rna.2039610. Epub 2010 Apr 29. RNA. 2010. PMID: 20430859 Free PMC article.
-
What Does the Future Hold for Yellow Fever Virus? (II).Genes (Basel). 2018 Aug 21;9(9):425. doi: 10.3390/genes9090425. Genes (Basel). 2018. PMID: 30134625 Free PMC article. Review.
-
Combinatorial Minigenome Systems for Emerging Banyangviruses Reveal Viral Reassortment Potential and Importance of a Protruding Nucleotide in Genome "Panhandle" for Promoter Activity and Reassortment.Front Microbiol. 2020 Apr 8;11:599. doi: 10.3389/fmicb.2020.00599. eCollection 2020. Front Microbiol. 2020. PMID: 32322247 Free PMC article.
References
-
- Brinton M A, Dispoto J H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162:290–299. - PubMed
-
- Brinton M A, Fernandez A V, Dispoto J H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. - PubMed
-
- Cahour A, Pletnev A, Vazielle-Falcoz M, Rosen L, Lai C J. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology. 1995;207:68–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

