Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes
- PMID: 11390596
- PMCID: PMC114310
- DOI: 10.1128/JVI.75.13.5949-5957.2001
Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes
Abstract
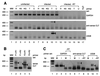
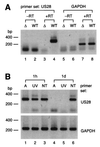
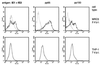
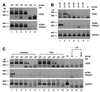
The human cytomegalovirus (HCMV) US28 gene product, pUS28, is a G protein-coupled receptor that interacts with both CC and CX(3)C chemokines. To date, the role of pUS28 in immune evasion and cell migration has been studied only in cell types that can establish productive HCMV infection. We show that HCMV can latently infect THP-1 monocytes and that during latency US28 is transcribed. We also show that the transcription is sustained during differentiation of the THP-1 monocytes. Since cells expressing pUS28 were previously shown to adhere to immobilized CX(3)C chemokines (C. A. Haskell, M. D. Cleary, and I. F. Charo, J. Biol. Chem. 275:34183-34189, 2000), we hypothesize that latently infected circulating monocytes express pUS28, thereby enabling adhesion of these cells to CX(3)C-exposing endothelium. Consequently, the US28-encoded chemokine receptor may play an important role in dissemination of latent HCMV.
Figures

Similar articles
-
Constitutive inositol phosphate formation in cytomegalovirus-infected human fibroblasts is due to expression of the chemokine receptor homologue pUS28.J Virol. 2003 Apr;77(8):4489-501. doi: 10.1128/jvi.77.8.4489-4501.2003. J Virol. 2003. PMID: 12663756 Free PMC article.
-
Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection.mBio. 2017 Dec 5;8(6):e01754-17. doi: 10.1128/mBio.01754-17. mBio. 2017. PMID: 29208743 Free PMC article.
-
The human cytomegalovirus-encoded chemokine receptor US28 induces caspase-dependent apoptosis.FEBS J. 2005 Aug;272(16):4163-77. doi: 10.1111/j.1742-4658.2005.04829.x. FEBS J. 2005. PMID: 16098198
-
US28 actions in HCMV infection: lessons from a versatile hijacker.Rev Med Virol. 2005 Jul-Aug;15(4):269-82. doi: 10.1002/rmv.468. Rev Med Virol. 2005. PMID: 15861487 Review.
-
US28: HCMV's Swiss Army Knife.Viruses. 2018 Aug 20;10(8):445. doi: 10.3390/v10080445. Viruses. 2018. PMID: 30127279 Free PMC article. Review.
Cited by
-
Identification of a novel signaling complex containing host chemokine receptor CXCR4, Interleukin-10 receptor, and human cytomegalovirus US27.Virology. 2020 Sep;548:49-58. doi: 10.1016/j.virol.2020.06.006. Epub 2020 Jun 17. Virology. 2020. PMID: 32838946 Free PMC article.
-
EBV BILF1 evolved to downregulate cell surface display of a wide range of HLA class I molecules through their cytoplasmic tail.J Immunol. 2013 Feb 15;190(4):1672-84. doi: 10.4049/jimmunol.1102462. Epub 2013 Jan 11. J Immunol. 2013. PMID: 23315076 Free PMC article.
-
Human Cytomegalovirus Interleukin 10 Homologs: Facing the Immune System.Front Cell Infect Microbiol. 2020 Jun 9;10:245. doi: 10.3389/fcimb.2020.00245. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32582563 Free PMC article. Review.
-
Control of Cytokines in Latent Cytomegalovirus Infection.Pathogens. 2020 Oct 21;9(10):858. doi: 10.3390/pathogens9100858. Pathogens. 2020. PMID: 33096622 Free PMC article. Review.
-
Tumor Necrosis Factor Alpha Induces Reactivation of Human Cytomegalovirus Independently of Myeloid Cell Differentiation following Posttranscriptional Establishment of Latency.mBio. 2018 Sep 11;9(5):e01560-18. doi: 10.1128/mBio.01560-18. mBio. 2018. PMID: 30206173 Free PMC article.
References
-
- Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greaves D R, Zlotnik A, Schall T J. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. - PubMed
-
- Billstrom M A, Lehman L A, Worthen G S. Depletion of extracellular RANTES during human cytomegalovirus infection of endothelial cells. Am J Respir Cell Mol Biol. 1999;21:163–167. - PubMed
-
- Bodaghi B, Jones T R, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier J L, Michelson S. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med. 1998;188:855–866. - PMC - PubMed
-
- Bolovan-Fritts C A, Mocarski E S, Wiedeman J A. Peripheral blood CD14+ cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93:394–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

